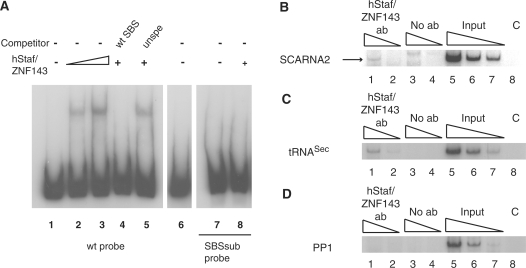
Figure 3.
Identification of an occupied hStaf/ZNF143 transcription factor binding site in the human SCARNA2 promoter. (A) Gel retardation assays were performed with 32P-labeled promoter fragments encompassing positions −88/+145 of the SCARNA2 promoter and containing the wild-type SBS sequence (wt probe, lanes 1–6), mutant SBS (SBSsub probe, lanes 7 and 8). In the SBSsub probe, the −70 CTCCCA−65 sequence was replaced by GATATC. Probes were incubated in the absence (lanes 1 and 7) or presence of hStaf/ZNF143 (2.5 µl of programmed reticulocyte lysate in lanes 2, 4, 5 and 8; 5 µl in lane 3). Reactions in lanes 4 and 5 were performed in the presence of a 1000-fold molar excess of unlabeled specific competitor (wt SBS) and unspecific competitor (unspe). Reaction in lane 6 was performed in the presence of 5 µl of unprogrammed reticulocyte lysate. Lanes 1–6 and 7–8 are from separate experiments. (B–D) Immunoprecipitation with hStaf/ZNF143 antibody of the SCARNA2 promoter from formaldehyde cross-linked chromatin prepared from HeLa cells. DNA fragments recovered from input chromatin, or ChIP assays with anti-hStaf/ZNF143 or control experiment without antibody, were analyzed by semi-quantitative PCR with specific primer pairs in the presence of (α−32P) dCTP. Lanes 1, 2 and 3, 4: serial dilutions of DNA immunoprecipitated with anti-hStaf/ZNF143 or no antibody, respectively. Lanes 5–7: serial dilutions of input material to demonstrate that the assays were within the linear range of PCR amplification. Lane 8 (C, PCR control): PCR lacking DNA template. (B) PCR was performed with specific primer pairs for the human SCARNA2 promoter or (C) for the tRNASec promoter acting as the positive control. (D) Negative control with the PP1 primer pair targeting a region located upstream of the tRNASec gene and lacking SBS.