Abstract
Actin depolymerizing factors (ADF/cofilin) modulate the rate of actin filament turnover, networking cellular signals into cytoskeletal-dependent developmental pathways. Plant and animal genomes encode families of diverse ancient ADF isovariants. One weakly but ubiquitously expressed member of the Arabidopsis ADF gene family, ADF9, is moderately expressed in the shoot apical meristem (SAM). Mutant alleles adf9-1 and adf9-2 showed a 95% and 50% reduction in transcript levels, respectively. Compared to wild-type, mutant seedlings and plants were significantly smaller and adult mutant plants had decreased numbers of lateral branches and a reduced ability to form callus. The mutants flowered very early during long-day light cycles, but not during short days. adf9-1 showed a several-fold lower expression of FLOWERING LOCUS C (FLC), a master repressor of the transition to flowering, and increased expression of CONSTANS, an activator of flowering. Transgenic ADF9 expression complemented both developmental and gene expression phenotypes. FLC chromatin from adf9-1 plants contained reduced levels of histone H3 lysine 4 trimethylation and lysine 9 and 14 acetylation, as well as increased nucleosome occupancy consistent with a less active chromatin state. We propose that ADF9 networks both cytoplasmic and nuclear processes within the SAM to control multicellular development.
Keywords: ADF, Cofilin, Epigenetic, Nuclear, Flowering time, Shoot apical meristem
Introduction
The actin-based cytoskeleton plays an essential role in plant and animal development (Jacinto and Baum 2003; Mathur 2004; Pawloski et al. 2006). Actin filaments and bundles dynamically contribute to the programming of organ, tissue, and cell development by facilitating the assembly of various cellular structures. Actin’s role in cell polarity and division plane determination is particularly important in plants, where cells do not migrate during multicellular organ development. Actin also has alternative nuclear roles controlling gene expression at the levels of chromatin remodeling and transcription (Bettinger et al. 2004;Miralles and Visa 2006; Visa 2005). Control of actin function is complicated by the expression of hundreds actin-binding proteins (ABPs) (Meagher and Fechheimer 2003), some of which participate in actin’s nuclear activities (Bettinger et al. 2004).
The actin depolymerizing factors (ADFs) and closely related cofilins in vertebrates and yeast are among the most highly expressed ABPs that regulate actin dynamics. In the cytoplasm, ADFs bind F-actin to alter its helical twist and bind actin monomers in response to diverse stimuli, with the overall effects of enhancing actin filament turnover and actin filament assembly (Bamburg 1999). ADFs and Cofilins also can be found in the nucleus and participate in the nuclear import of actin (Bamburg 1999; Ruzicka et al. 2007). Via their role of shuttling actin into the nucleus, ADFs may participate in the chromatin-level control of gene expression and perhaps in the epigenetic determination of cell fate. A theme emerging from data on nuclear activities for actin and ABPs like ADF is that there may be essential “crosstalk” between the nuclear and cytoplasmic compartments (Minakhina et al. 2005). This communication could provide both compartments with important dynamic cues as to the status of expanding and dividing cells and developing organs.
In both plants and mammals the ADFs are encoded by an ancient gene family. The Arabidopsis (Arabidopsis thaliana) ADF family is comprised of 11 gene members in several divergent classes (Maciver and Hussey 2002; Ruzicka et al. 2007). Arabidopsis ADF9 (At4g34970) is weakly expressed in nearly all vegetative tissues, but is more strongly expressed in shoot apical meristem (SAM) and root sub-apical region, trichomes, and callus (Ruzicka et al. 2007). Herein, we have characterized strong and weak knockdown alleles of ADF9 and shown the defective plants had dramatic alterations in multicellular development. Most of the observed phenotypes might be ascribed to altered cytoplasmic cytoskeletal activities affecting the meristem and shoot development. However, both alleles flowered early relative to the wild-type Arabidopsis thaliana Columbia ecotype and transcripts for transcription factors known to control the transition to flowering in the apical meristem, such as FLOWERING LOCUS C (FLC) and CONSTANS (CO) (Michaels and Amasino 1999; Putterill et al. 1995), were significantly mis-regulated in adf9-1. The structure of chromatin at the FLC locus was in a more repressed state in adf9-1, in terms of both histone modification and nucleosome occupancy. Our data suggest that ADF9 has both cytoplasmic and nuclear activities affecting multicellular development.
Materials and methods
Plant materials and growth conditions
Mutants in the ADF9 gene (At4g34970) were obtained from TAIR (www.arabidopsis.org) adf9-1 (SALK_056064) and adf9-2 (#Garlic_760_A03.b.1.a.Lb3Fa). Both T-DNA insertion mutants were in an Arabidopsis thaliana Columbia ecotype genetic background. ADF9 is so named in Ruzicka et al (2007) and named “similar to ADF5” (AT2G16700) at TAIR. The mutants were backcrossed twice with wild-type Columbia and examined for a single insertion before phenotypic analysis. Experiments were performed on T4 or T5 generation selfed seedlings or plants. All controls were performed with the Columbia ecotype. Seeds from wild-type, adf9-1, and adf9-2 plants were sown directly into soil (Fafard). After 2 days at 4°C, they were moved to growth chambers (t = 0) maintained at 22°C under long-day (16 h light and 8 h dark) or short-day (9 h light and 15 h dark) photoperiod. Tissues were harvested 3 h after the beginning of the light cycle. Ten days after germination, the seedlings were transferred to 2.5 in. pots and grown under the same conditions. For experiments on seedlings seeds were sterilized and plated on MS phytoagar (Murashige and Skoog 1962) supplemented with 1% sucrose, stratified at 4°C for 2 days, and grown at 22°C (t = 0) for 12 days.
Determination of genotypes by PCR
Allele and gene specific primers were designed to amplify identifying fragments from the wild-type ADF9, adf9-1 or adf9-2 alleles. A 767 bp fragment for the ADF9 allele was amplified using the sense primer ADF9-5′utrS (5′-GAAAATATTTTGGATGATTGGTATATA) and antisense primer ADF9-3′utrA (5′-ACGATATAACTCCAGTTATGTGTTGTGA). A 401-bp fragment of the adf9-1 mutant allele was amplified with a left border T-DNA primer LBaI (5′-TGGTTCACGTAGTGGGCCATCG) and the antisense primer ADF9-3′urtA. A 366 bp fragment of the adf9-2 mutant allele was amplified with a left border T-DNA primer LB3 (5′-TAGCATCTGAATTTCATAACC) and the sense primer ADF9-5′urtS. The PCR products of both mutant alleles were sequenced to confirm the exact location of the T-DNA insertion. Homozygous mutant plants derived from two backcrosses to wild-type were genotyped by PCR to verify the correspondence of homozygous genotype with various phenotypes. The DNA used as a template for genotype and sequence determination was prepared by a modified rapid alkali DNA screening method (Gilliland et al. 1998) or with CTAB according to Doyle et al. (1990).
Flowering time and inflorescence phenotypes
The total number of rosette leaves at the time in which the first inflorescence could be distinguished from leaf primordia was recorded as a measure of flowering time. At least 24 plants were assayed to determine the flowering time and other phenotypes of each plant line, with four biological replicates to confirm the observations. Nine days after flowering the length of the secondary branches and the number of primary inflorescences was recorded. Gene expression assays to examine the flowering time signaling pathway were performed on 10-day-old seedlings germinated and grown vertically on MS agar.
Mutant complementation and ADF9 overexpression
Complementation and overexpression studies were carried out by first cloning the 426 bp ADF9 cDNA protein encoding sequence into the NcoI/BamHI replacement region of actin ACT2 promoter and terminator expression cassette pA2pt (Kim et al. 2005). An NcoI site containing the ATG start codon and a BamHI site following the stop codon were introduced to the ADF9 coding sequence by mutagenic PCR. The fusion between the expression cassette and ADF9 was subcloned into the pCambia binary vector (Hajdukiewicz et al. 1994) using the flanking KpnI and SacI sites to make pA2pt::ADF9 in E. coli. pA2pt::ADF9 was transformed into the Agrobactirium strain C58C1. Plants were transformed by Agrobacterium-mediated vacuum infiltration method (Bechtold and Pelletier 1998). Complemented plants were selected by plating the seeds on germination medium (0.5× MS, 1% sucrose, phytagar germination medium containing 50 mg−1 hygromycin) and after 3 days transferred to soil. Multiple independent T2 lines were generated and compared to adf9-1 and wild-type seedlings and plants without drug selection.
GFP reporter for F-actin
We transformed wild-type and adf9-1 mutant plants with the F-actin reporter ABD2-GFP (Wang et al. 2004). Multiple lines of positive transformants were selected on hygromycin and vertically grown on 0.5 MS agar plates. Microfilaments were visualized in trichomes as described previously (Wang et al. 2004) using a Leica confocal laser-scanning microscope (TCS-SP2, Heidelberg, Germany).
Quantitative Real Time PCR analyses
RNA was isolated from 10-day-old seedlings using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA), and 1.5 µg of total RNA from each sample was transcribed into cDNA with the Super Script III kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions except that incubations were performed for only 30 min and at 55°C using oligo (dT) primer. Aliquots of the cDNA were used as template for the qRT-PCR analyses in triplicate reactions for each of three biological replicates on an Applied Biosystems 7500 Real Time PCR Instrument. Real time PCR reactions consisted of SYBR GREEN PCR Master Mix (Applied Biosystems, Foster City, CA, USA), 0.4 µM of each primer, 1:50 dilution of cDNA in a 25 µl reaction volume. Primer sets used for qRT-PCR assays of transcript levels are as follows: ADF9 (ADF9-RTS: 5′-ATATAACGAAAGAACAAGAAGACA-3′, ADF9-RTA: 5′-CACTCGTCGCCGTCTTCAA-3′); 18S rRNA (18 s-RT2S: 5′-GGGGGCAATCGTATTTCATA-3′, 18S-RT2A: 5′-TTCGCAGTTGTTCGTCTTTC-3′); ACT2 (ACT2-RTS: 5′-GATGAGGCAGGTCCAGGAATC-3′, ACT2-RTA: 5′-AACCCCAGCTTTTTAAGCCTTT-3′); UBQ10 (UBQ-RTS: 5′-AGAAGTTCAATGTTTCGTTTCATGTAA-3′, UBQ-RTA: 5′-GAACGGAAACATAGTAGAACACTTATTCA-3′); FLC (FLC-RTS: 5′-CATCATGTGGGAGCAGAAGCT-3′, FLC-RTA: 5′-CGGAAGATTGTCGGAGATTTG-3′); CO (CO-RTS: 5′-TGGCAAAACTAGACTGCATGCT-3′, CO-RTA: 5′-CCCTATATGCATAAAACCGTGGTAA-3′); FT (FT-RTS: 5′-GGCGCCAGAACTTCAACACT-3′, FT-RTA: 5′-CGGGAAGGCCGAGATTG-3′); SOC1 (SOC1-RTS: 5′-AAAGCTCTAGCTGCAGAAAACGA-3′, SOC1-RTA: 5-GACCAAACTTCGCTTTCATGAGAT-3′); LFY (LFY-RTS: 5′-TTGTCGTCATGGCTGGGATATA-3′, LFY-RTA: 5′-GAACATACCAAATAGAGAGACGAGGAT-3′); and AP1 (AP1-RT-S = 5′-CGCAGCAGC ACCAAATCC-3′. AP1-RT-A 5′-TGAGAAAAGGAGATGGCTGATG-3′). We used the 2−ddCt method (Livak and Schmittgen 2001) to determine Relative Quantification values.
Chromatin structure assays at FLC
ChIP assays on H2A.Z deposition at the FLC locus were performed on sheared chromatin from 10-day-old shoots grown under long day conditions as described previously (Deal et al. 2007) using the same first three sets of FLC PCR primers (FLC1, 2, 3). ChIP assays for histone H3 trimethylation at lysine 4 and H3 acetylation at lysine 9 and 14 were performed identically except that the H3K4me3 (#07–473) and H3K9K14ac (#06–599) antibodies from Upstate (Lake Placid, NY) were used in the precipitation assays. The first two ChIP assays were normalized to a region in the ACT2 3′UTR and the H3K9K14 assays were normalized to a LINE2 element. The three primer pairs were describe previously (Deal et al. 2007).
Nucleosome occupancy was assayed in the promoter region of FLC by a modification of the method described for yeast genes by Sekinger et al. (2005) starting with 1 g of 10-day-old whole seedlings grown under long-day length conditions. Nucleosomes were prepared by a modification of an older protocol (Vega-Palas and Ferl 1995). The qPCR assays on nucleosomal DNA were performed using the primer pairs listed in Table 1. Relative quantity (RQ) data were normalized first to input DNA concentration by subtracting the raw Cycle Threshold (CT) value for actin 2 (ACT2) from the Target CT value and second the results were normalized to the efficiency of PCR amplification of each primer pair performed on purified Arabidopsis DNA. This latter efficiency of DNA amplification on naked DNA varied by only two-fold among the primer pairs used in this study. qPCR was performed as above for qRT-PCR. The nucleosome preparation, qPCR assays, and calculations are detailed in the Supplemental Section.
Table 1.
Primers for FLC nucleosome occupancy assay
| Name | Sequence |
|---|---|
| FLC-nuc-P1S | 5′-CAAGCTGATACAAGCAAAAAAGAA-3′ |
| FLC-nuc-P1N | 5′-AATACAAGAAATCTTAAATGTCCACAC-3′ |
| FLC-nuc-P2S | 5′-GTGTGGACATTTAAGATTTCTTGTATT-3′ |
| FLC-nuc-P2N | 5′-CGGGAGATTAACACAAATAATAAAGG-3′ |
| FLC-nuc-P3S | 5′-CCTTTATTATTTGTGTTAATCTCCCG-3′ |
| FLC-nuc-P3N | 5′-CGCCTACGTCATCAAATTTTATAAA-3′ |
| FLC-nuc-P4S | 5′-TTTATAAAATTTGATGACGTAGGCG-3′ |
| FLC-nuc-P4N | 5′-ATTTGGCAGTTAATTAGTAGGTGTT-3′ |
| FLC-nuc-P5S | 5′-AACACCTACTAATTAACTGCCAAAT-3′ |
| FLC-nuc-P5N | 5′-ATATCGAAACCTAAAACTGGTTTGA-3′ |
| FLC-nuc-P6S | 5′-TCAAACCAGTTTTAGGTTTCGATAT-3′ |
| FLC-nuc-P6N | 5′-CTATTGCAGAAAGAACCTCCA-3′ |
| FLC-nuc-P7S | 5′-TGGAGGTTCTTTCTGCAATAG-3′ |
| FLC-nuc-P7N | 5′-GTTTTCACTTTCGTTGCTAAATGAA-3′ |
| FLC-nuc-P8S | 5′-TTCATTTAGCAACGAAAGTGAAAAC-3′ |
| FLC-nuc-P8N | 5′-TTGTGTTTTGAAGACAAGATTGC-3′ |
| FLC-nuc-P9S | 5′-GCAATCTTGTCTTCAAAACACAA-3′ |
| FLC-nuc-P9N | 5′-TTTTTTTTTTTTGGGGGTAAACGA-3′ |
| FLC-nuc-P10S | 5′-TCGTTTACCCCCAAAAAAAAAAAA-3′ |
| FLC-nuc-P10N | 5′-TTCTTTCTATTTTTTGTGCCTATCTAC-3′ |
| FLC-nuc-P11S | 5′-GTAGATAGGCACAAAAAATAGAAAGAA-3′ |
| FLC-nuc-P11N | 5′-GAGATACTAAGCGTTTTCTCTTTCTA-3′ |
| FLC-nuc-P12S | 5′-TAGAAAGAGAAAACGCTTAGTATCTC-3′ |
| FLC-nuc-P12N | 5′-TTTGTGCCCTAATTTGATCCT-3′ |
| FLC-nuc-P13S | 5′-AGGATCAAATTAGGGCACAAA-3′ |
| FLC-nuc-P13N | 5′-TCAATTCGCTTGATTTCTAGTTTTTT-3′ |
| FLC-nuc-P14S | 5′-AAAAAACTAGAAATCAAGCGAATTGA-3′ |
| FLC-nuc-P14N | 5′-CTTTCTCGATGAGACCGTT-3′ |
Assays of GUS after treatment with various phytohormones
An ADF9 promoter-β-glucuronidase (GUS) reporter fusion (ADF9pt::GUS) was transformed into wild-type Columbia and a single insertion homozygous line generated by selfing as described previously (Ruzicka et al. 2007). This line was subjected to treatment with various phytohormones and assayed for GUS enzyme expression as modified from Jefferson et al. (1987). Seedlings were transferred from vertically grown plates to soft agar plates plus hormones and allowed to grow for an additional 20 h. Hormone stocks were each prepared at 100 mM in DMSO. GUS expression was assayed by incubating seedlings in 50 mM NaPO4 pH 7.0, containing 0.5% Triton X-100, 0.5 mM X-gluc (Jersey Lab Supply, Livingston, NJ), 0.5 mM K4(Fe(CN)6) for 45 min at 37°C. Seedlings were removed from solution and bleached in successive washes of 70% ethanol before being photographed using a Leica dissecting microscope (Leica Microsystems) equipped with a Hamamatsu CCD camera.
Assay of callus formation
Wild-type and adf9-1 seeds were plated on MS media and grown vertically for 2 weeks. Ten leaves (1.5 cm) and 10 roots (2.5 cm long sections) of equal size were collected for both wild-type and adf9-1 seedlings and moved to callus-inducing media supplemented with 4.5 µM 2,4-D and 25 nM kinetin (Kandasamy et al. 2001). The plates were photographed at time zero (not shown), wrapped in foil, and callus was allowed to grow for 2 and 4 weeks. At 2 and 4 weeks the foil was removed, photographs were taken, and callus fresh weight was measured.
Results
ADF9-defective alleles
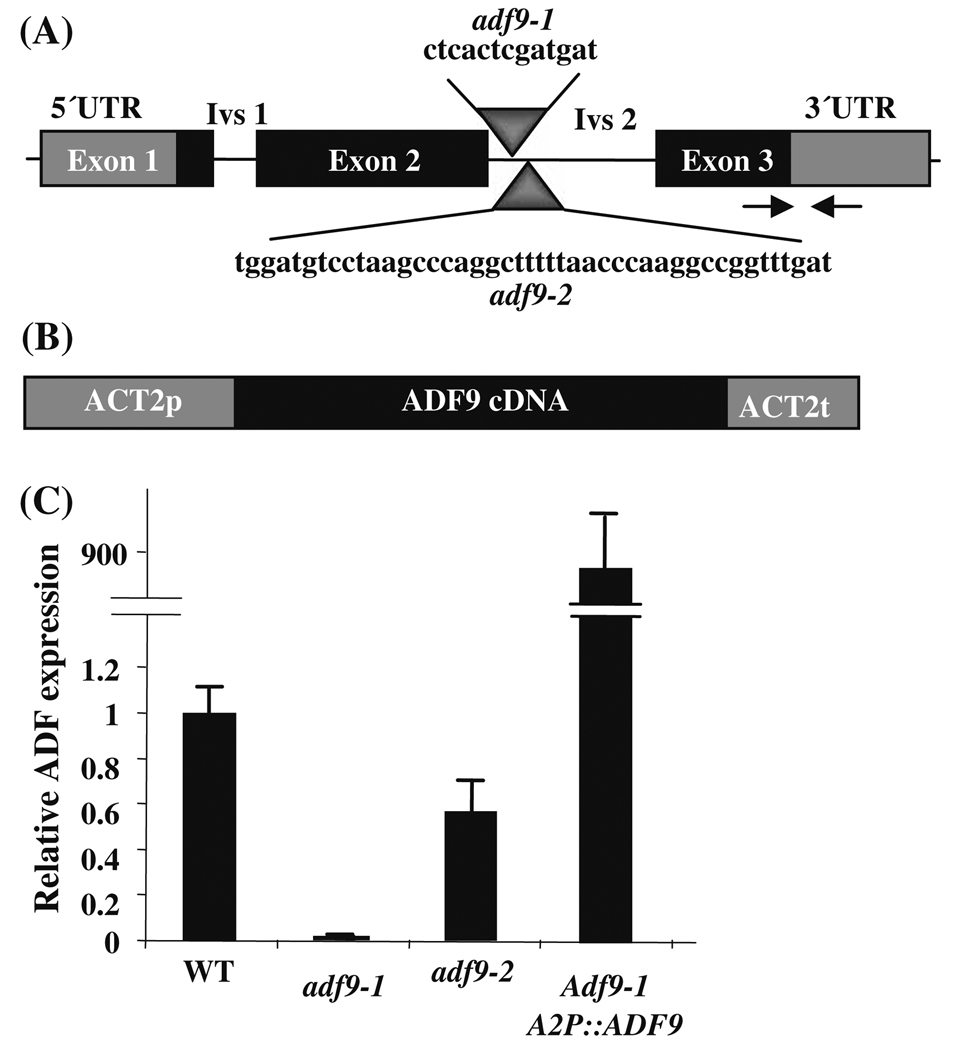
Two independent ADF9 insertion mutant alleles, adf9-1 and adf9-2, were isolated in the Arabidopsis Columbia ecotype. They each contained a T-DNA insertion that mapped within the second intron as diagrammed in Fig. 1. The insertion in adf9-1 was located 78 bp downstream from the splice donor site and deleted 13 bp of downstream sequences from intron 2. The insertion in adf9-2 was located 114 bp from the splice donor site and deleted 44 bp from intron 2. Hence, the deleted regions in adf9-1 and adf9-2 do not overlap. ADF9 mRNA levels were assayed in the two mutants relative to the levels of 18S rRNA and normalized to wild-type in multiple samples of 10-day-old seedlings using quantitative Real Time Polymerase Chain Reaction (qRT-PCR). ADF9 transcript levels were 95 to 99% reduced in adf9-1, but only 50% reduced in adf9-2. Because the ADF9-specific primer pair used to assay mRNA levels was positioned to span the first intron (Fig. 1a) and was located upstream of both T-DNA insertions these data must reflect a lowered expression and/or lowered stability for the truncated transcripts. Furthermore, little truncated ADF9 protein could be produced in the adf9-1 allele. Similar, reductions in transcript levels were observed using other ADF9-specific primer pairs located downstream of these insertions during qRT-PCR. Thus, adf9-1 is essentially a null allele, while adf9-2 is a weak knockdown allele. Similar reductions in relative ADF9 transcript levels were obtained when these qRT-PCR data were normalized to total input RNA or to actin ACT2 or ubiquitin UBQ10 mRNAs instead of 18S rRNA.
Fig. 1. Gene constructs and reduced mRNA expression in adf9-1 and adf9-2 mutants.
(a) The T-DNA insertions in adf9-1 and adf9-2 were both located within intron 2, were separated at their 5′ ends by 37 bp, and were not overlapping. The intron sequences deleted from each intron and replaced by T-DNA are shown for each insertion. Arrows indicate the location of the qRT-PCR primers used to assay ADF9 transcript levels. (b) The cDNA encoding region of ADF9 was cloned within the actin ACT2 constitutive expression cassette A2pt. A2pt::ADF9 was used to complement adf9-1. (c) The adf9-1 and adf9-2 mutations produced a 95% and 50% drop in levels of transcript relative to wild-type, respectively. The adf9-1 insertion may disrupt an important transcriptional enhancer or splicing sequence resulting in lower steady state mRNA levels. Complementing adf9-1 with A2pt::ADF9 resulted in an approximately 900-fold increase in ADF9 transcript levels over wild-type
Developmental phenotypes of ADF9-defective alleles
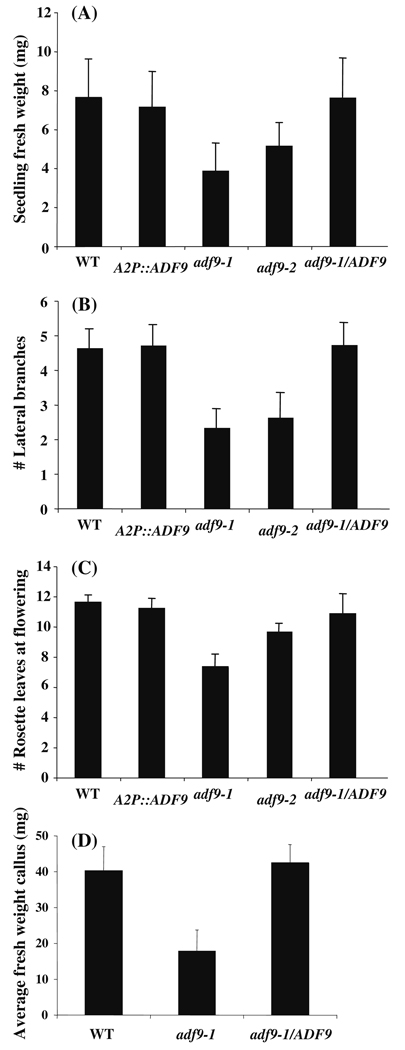
The adf9-1 and adf9-2 alleles displayed similar pleiotropic defects in growth and development as shown in Fig. 2. Twelve days following germination, homozygous mutant seedlings of the two alleles were reduced 2-fold and 1.5-fold in size and fresh weight, respectively, in comparison to wild-type, as quantified in Fig. 3. The adf9-1 and adf9-2 alleles flowered approximately 16 and 18 days after germination, respectively, compared to 22 days for wild-type Columbia controls that were germinated on soil and grown under long-day conditions: 16 h of light and 8 h of darkness. For the null allele, adf9-1, the inflorescence first emerged with only 7 rosette leaves compared to 10 leaves for adf9-2 and 12 leaves for wild-type Columbia. For the adf9-1 allele, the accelerated transition to flowering resulted in slightly dwarfed adult plants. Surprisingly, no significant differences in seed set were observed in the mutants relative to wild-type. When adf9-1 plants were grown under short-day conditions, 9 h of light and 15 h of darkness, they flowered at the same time as wild-type (Fig. 2d). Therefore, the defect in flowering time transition may be photoperiod dependent. Both mutants showed increased apical dominance, revealed by fewer lateral branches than wild-type assayed 9 days after bolting (Fig. 2b and Fig. 3b). For example, the 25-day-old adf9-1 plants generally had two to three lateral branches, while 31-day-old wild-type plants of about the same developmental stage had four to five. Although the adf9 defective lines had fewer lateral branches, the length of individual lateral branches was longer, such that the total length of all lateral branches was indistinguishable between mutants and wild-type (not shown).
Fig. 2. Morphological phenotypes for adf9-1 and adf9-2.
(a) Twelve-day-old adf9-1 and adf9-2 mutant seedlings are smaller compared to wild-type. (b) Mutants have an increase of apical dominance in the inflorescence as revealed by a decrease number of secondary branches and longer branches compared to wild-type. (c) adf9-1 and adf9-2 mutants flower earlier with fewer rosette leaves than wild-type under long-day growth. (d) adf9-1 flowers at approximately same time as wild-type under short-day growth conditions
Fig. 3. Quantification of adf9-1 and adf9-2 mutant phenotypes and complemented lines.
(a) The weight of 10-day-old adf9-1 and adf9-2 mutant seedlings was compared to wild-type and adf9-1 mutant line complemented with the cDNA expression clone A2p::ADF9. (b) A comparison was made of the number of secondary branches and longer branches compared to wild-type and adf9-1 complemented with the ADF9 cDNA. (c) Early flowering was assayed as the number of rosette leaves at the time the inflorescence first emerges is compared. (d) The average fresh weight of callus was measured after two weeks on callus induction media. Ten or more seedlings or plants or root callus samples were measured for each assay. Error bars represent the standard error from the mean
Genetic complementation and overexpression
A2pt::ADF9, a construct overexpressing ADF9 cDNA from a strong constitutive actin ACT2 expression cassette (Fig. 1b) was introduced into adf9-1 plants. This complementation experiment was performed to confirm that the phenotypes observed for the adf9-1 allele were due entirely to the disruption of the ADF9 gene and test the effect of overexpressing ADF9. The A2pt::ADF9 construct quantitatively complemented all adf9-1 morphological phenotypes. Seedling size, apical dominance, and flowering time were indistinguishable from wild-type in the complemented lines as quantified for a single representative line in Fig. 3. The A2pt::ADF9 transgene produced extremely high levels of the ADF9 transcript, often several hundred-fold higher than wild-type as shown for one line (Fig. 1c). This high expression is consistent with the high levels of expression achieved with the A2pt expression cassette in previous studies (Li et al. 2005). This strong overexpression of ADF9 mRNA did not produce dominant morphological phenotypes that differed from wild-type when expressed in the adf9-1 or wild-type backgrounds (Fig. 3).
ADF9 is a repressor of flowering
The early flowering phenotypes of the adf9-1 and adf9-2 mutants partially resembled those of several chromatin remodeling mutants negatively affecting transcript levels of FLOWERING LOCUS C (FLC) (He and Amasino 2005) or positively affecting CONSTANS (CO) (Gaudin et al. 2001). Therefore, we considered the possibility that the early flowering of ADF9-defective plants were due to changes in gene expression and not directly due to defects in the F-actin cytoskeleton. FLC is a master repressor of the transition from vegetative to reproductive growth in Arabidopsis. FLC represses FLOWERING LOCUS T (FT) and SUPPRESSOR OF CONSTANS1 (SOC1), which function to activate APETLA1 (AP1) and LEAFY (LFY), and advance flowering, as summarized in Fig. 4. We reasoned that appropriate levels of ADF9 protein might be required to shuttle actin into the nucleus, where actin participates in chromatin remodeling complexes or transcriptional machinery that control the expression of flowering time regulators, like FLC. Alternatively, ADF9 might have unknown actin-independent roles in regulating gene expression.
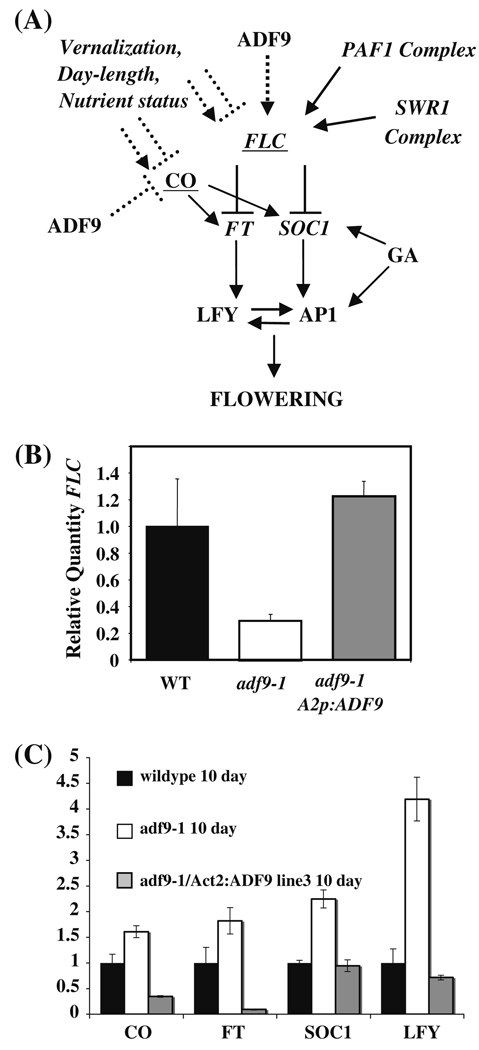
Fig. 4. ADF9 alters expression of genes controlling flowering time.
(a) Model for the signaling that occurs in the transition to flowering and the role of ADF9. FLC is a central repressor of this transition controlled by numerous environmental cues and factors that act via diverse changes in chromatin structure at the FLC locus. (b) The adf9-1 line had decreased levels of transcript encoding FLC relative to wild-type as assayed by qRT-PCR. FLC levels were restored in several independent lines complemented with A2pt::ADF9. (c) qRT-PCR assays of transcript levels for several other activators of flowering
To test the proposal that ADF9 plays a role in controlling gene expression, we examined the adf9-1 mutant and wild-type seedlings for the relative expression of FLC. We observed that FLC transcript levels in 10-day-old seedlings were four-fold lower in the mutant than wild-type (Fig. 4b). The relative quantities of transcripts in each sample were first measured with respect to 18S rRNA and then normalized to target transcript levels in wild-type. In addition, transcript levels from the downstream flowering activators, FT, SOC1, and LFY, were up-regulated 1.8-, 2- and 4-fold (Fig. 4c), respectively in adf9-1, consistent with loss of FLC expression (He and Amasino 2005). Mutant lines complemented by the overexpression of ADF9 cDNA (Fig. 3c) not only flowered at normal times, but they showed FLC transcript levels that were restored to those observed in wild-type controls and not significantly higher than wild-type as show for three complemented lines (Fig. 4b). Thus, the reduction in FLC levels and increases in the downstream flowering activators appeared to account for part of the early flowering phenotype of the adf9-1 mutant (He and Amasino 2005; Michaels and Amasino 1999).
Considering that ADF9-defective plants flowered early under long-day, but not short-day growth conditions, it was possible that ADF9 also acted in the photoperiod-dependent flowering pathway. The transcription factor CO activates FT and SOC in response to long-day growth conditions (Fig. 4a). Although there is considerably less evidence than for FLC, the CO locus also may be controlled at the level of chromatin remodeling (Gaudin et al. 2001). We observed a 1.6-fold increase in the levels of CO transcripts in 10-day-old adf9-1 seedlings (Fig. 4c). These data are consistent with ADF9 functioning as a repressor of CO and playing a role in the photoperiod-dependent flowering pathway.
Similar gene expression data to those presented herein were also obtained with comparisons of 12- and 14-day-old wild-type and adf9-1 seedlings. It should also be noted that an FLC null mutant, flc-3 (Michaels and Amasino 1999), flowered early under the long-day growth conditions we used in these experiments and we found that the lack of FRIGIDA expression in Columbia only partially attenuated the activity of FLC (Deal et al. 2007).
We examined A2pt::ADF9 complemented mutant lines for transcripts levels of those flowering time activators that were increased in the adf9-1 mutant as shown for one line in Fig. 4c. Complementation restored the levels of transcripts encoding the downstream activators, SOC1 and LFY, back down two wild-type levels. In contrast, transcript levels for the high level activators, CO and FT, were reduced significantly below wild-type levels.
ADF9 is essential to maintain normal chromatin structure at FLC
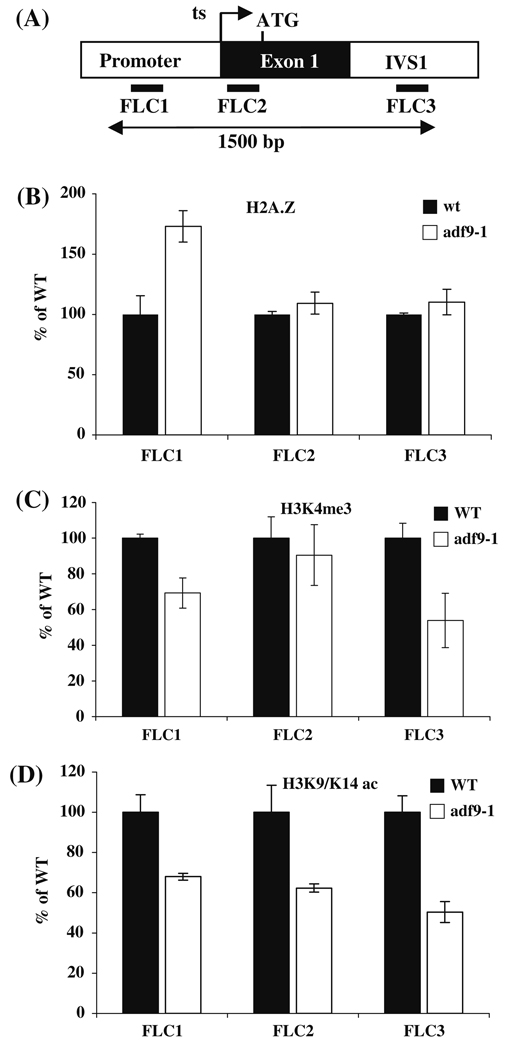
A number of defects in chromatin remodeling machinery cause the down-regulation of FLC gene expression (He and Amasino 2005). Therefore, it seemed reasonable to propose that the reductions in FLC expression in adf9-1 seedlings were due to the inability of ADF9 to contribute to normal chromatin remodeling at FLC. We performed a series of Chromatin Immuno-Precipitation (ChIP) assays to look for differences in histone modifications and histone variant subunits deposited in FLC chromatin, comparing wild-type and adf9-1 seedlings grown under long-day conditions. High levels of histone H2A.Z occupancy are observed at the 5′ and 3′ ends of the FLC transcript coding region, and mutations that cause significant loss of H2A.Z from these regions are associated with dramatically decreased FLC expression and early flowering in the Columbia ecotype (Deal et al. 2007). Decreased levels of histone H3 trimethylation at lysine 4 (H3K4me3) and H3 acetylation at lysine 9 and 14 (H3K9K14ac) flanking the promoter region of FLC are associated with lower levels of FLC expression (He and Amasino 2005; Martin-Trillo et al. 2006).
ChIP assays were performed on formalin fixed and sheared chromatin, using antibodies to H2A.Z, H3K4me3, and H3K9K14ac to enrich nucleosome bound DNA. We assayed three sites at the 5′ end of the FLC locus located in the promoter (FL1), the transcription start site (FL2), and first intron (FL3) by quantitative PCR (qPCR) analyses of the precipitated DNA, as shown in Fig. 5. H2A.Z deposition at the two downstream sites in FLC (Primer sets FLC2 and FLC3) was similar to wild-type, while H2A.Z occupancy in the upstream promoter site (Primer set FLC1) was increased 70% above wild-type (Fig. 5b). In the FLC promoter an inverse correlation has been reported between H2A.Z occupancy and gene expression in some genotypes and tissues via a mechanism discussed previously (Deal et al. 2007). A 30 to 45% drop in the levels of H3K4me3 was observed at two of the three sites in FLC (Primer sets FLC1 and FLC3) (Fig. 5c). Similarly, a 35 to 50% drop in the levels of H3K9K14ac was observed at all three sites in FLC in the adf9-1 mutants relative to wild-type (Fig. 5d).
Fig. 5. Chromatin remodeling phenotypes of adf9-1 at the FLC locus. ChIP assays were performed on sheared FLC chromatin.
(a) qPCR was used to amplify products within the promoter (FLC1), at the start of transcription (ts, FLC2), and in the first intron (ivs, FLC3). ChIP assays were performed to examine: (b) H2A.Z deposition; (c) histone H3 trimethylation at lysine 4 (H3K4Me3); and (d) H3 acetylation at lysine 9 and 14 (H3K9K14Ac)
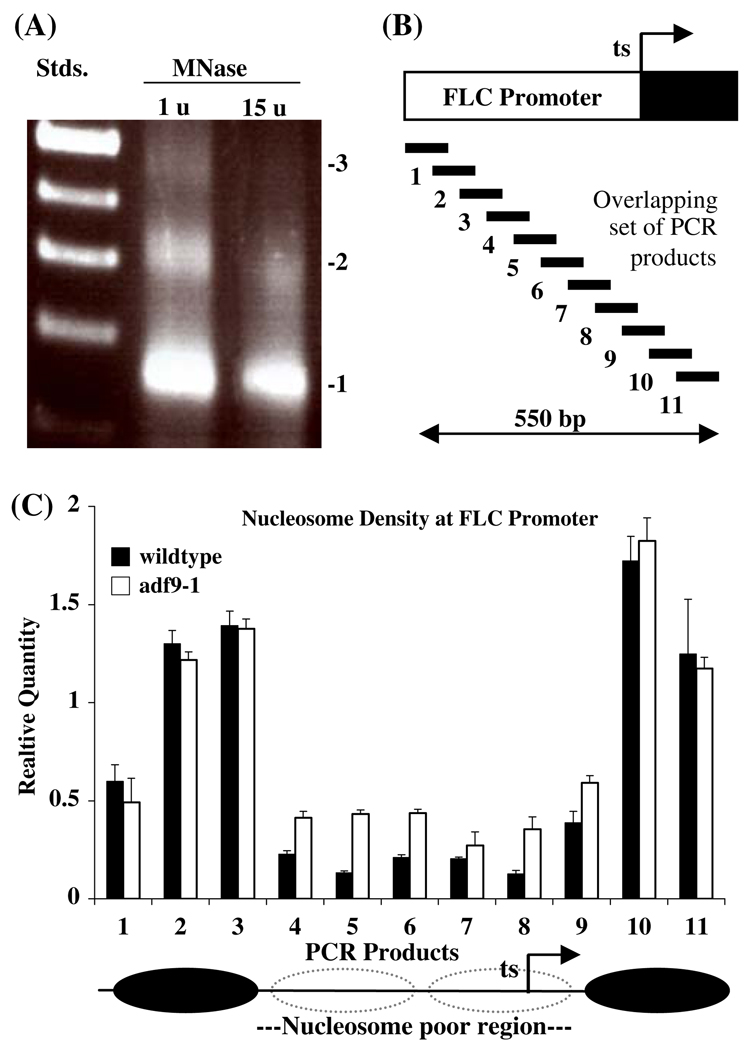
Increased nucleosome density, particularly in the promoter of a gene, is often associated with decreased rates of transcription. Therefore, we performed an assay of nucleosome occupancy within the FLC locus in adf9-1 and wild-type using qPCR. Nucleosomes prepared from 10-day-old Arabidopsis seedlings and digested with increasing levels of micrococcal nuclease contained DNA fragments of the sizes expected for tri- (450 bp), di- (~ 300 bp) and mononucleosomes (~ 150 bp) with a moderate nuclease concentration (Fig. 6). Primarily monosomal 150 bp DNA was observed at a 15-times higher nuclease concentration. Quantitative PCR primers were designed to amplify 11 nested products of approximately100 bp overlapping by approximately 50 bp from the monosomal preparation giving 50 bp resolution to the assay (Fig. 6b). The 550 bp region assayed covered the start of transcription (products 8, 9, 10, 11) and extended 350 bp upstream into the promoter. The nucleosome scanning assay of wild-type chromatin (15 u, Fig. 6a) revealed an approximately 300 bp nucleosome poor region (i.e., low product amplification) centered 100 bp upstream of the start of transcription (products 4 through 9, Fig. 6c). This region was flanked by two nucleosome protected regions that amplified efficiently (products 2, 3, 10, 11 in Fig. 6c). The adf9-1 mutant revealed 50 to 300% higher levels for five of the six FLC products amplified (e.g., products 4, 5, 6, 8, 9) in the nucleosome poor region relative to wild-type. Thus, nucleosome density in the nucleosome poor region appears much higher in the adf9-1 mutant than in wild-type.
Fig. 6. Nucleosome occupancy in the FLC promoter region.
(a) Arabidopsis nucleosomal DNA separated on an agarose gel and fluorescently stained with ethidium. Size standard ladder (Stds.); The two samples of DNA were prepared from nucleosomes digested with micrococcal nuclease (MNase) at 1 unit (u) and 15 units per 300 µl reaction. The position of mono- (1), di- (2), and trinucleosomes (3) are indicated. DNA from the 15 u digestion was examined in plate c. (b) Location of qPCR primers used to assay nucleosome protected DNA relative to a map of the FLC promoter region. (c) Nucleosome scanning assay the promoter region of FLC in wild-type and the adf9-1 mutant presented as the Relative Quantity of PCR amplification of the 11 products mapped in “a”. Standard errors are indicated for three replicate experiments. The normalized RQ values for purified gDNA = 1 for all products and are not shown. Possible nucleosome positions and the nucleosome poor region are indicated below the graph. qPCR products 8 and 9 and product 14 partially overlap with above qPCR products FLC1 and FLC2, respectively, in Fig. 5a
ADF9 expression is controlled by hormones
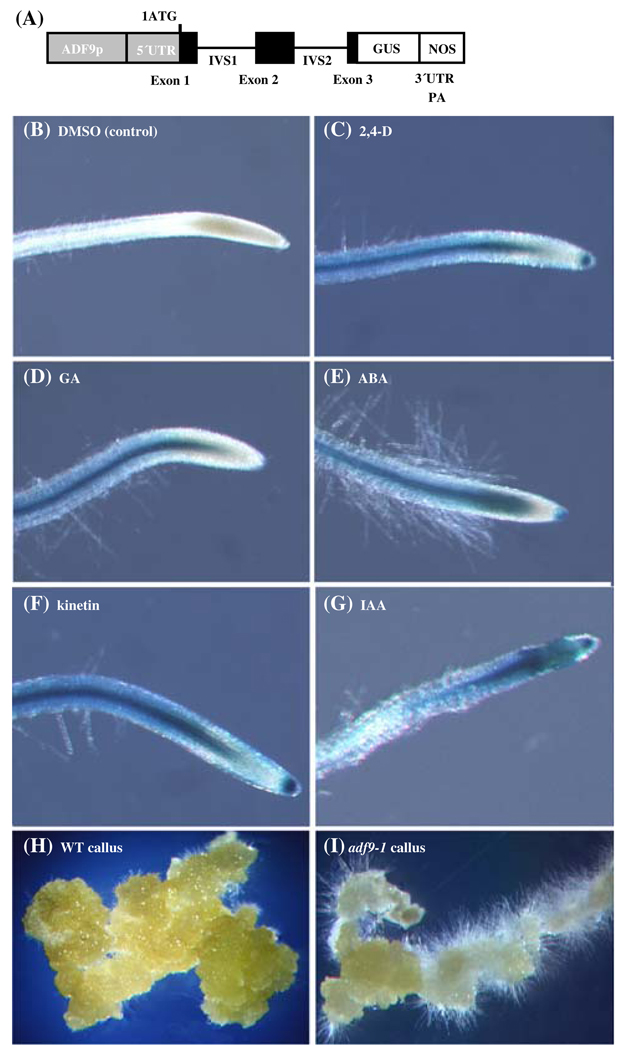
Phytohormones are implicated in diverse mechanisms of epigenetic control over organ initiation from the SAM and may affect flowering time (Hay and Tsiantis 2005; Veit 2006). Considering that ADF9 was required for the normal regulation of FLC transcript levels and may be acting through epigenetic mechanisms in the SAM, previous microarray data reporting that ADF9 transcript levels are dramatically increased by treatment with auxin (Zimmermann et al. 2004) may be relevant to the phenotypes observed for ADF9 defective plants. We used the ADF9p::GUS reporter construct described earlier (Ruzicka et al. 2007) to examine further the hormone induction of ADF9 in seedlings. ADF9p::GUS plants were transferred to growth media supplemented with various phytohormones for 20 h and then histochemically stained for GUS expression for 45 min. The ADF9p::GUS plants displayed an increase in GUS reporter expression in the SAM, trichomes, and root tip meristem in response to 2,4-dichlorodiphenoxyacetic acid (2,4-D) and indoleacetic acid (IAA), and a smaller increase in expression in these tissues in response to kinetin, gibberellic acid (GA3), and abscisic acid (ABA). The moderately high levels of activity of ADF9p::GUS in the SAM (Ruzicka et al. 2007) made hormonal induction over background difficult to visualize. Whereas basal ADF9p::GUS expression was low in roots, making increases in reporter expression easier to observe as shown in Fig. 7. All five hormone treatments produced a strong to moderate induction of the reporter in the sub-apical tissues of the root. These data suggest phytohormones play a role in the regulation of endogenous ADF9 levels and could contribute to an ADF9-dependent epigenetic control of development.
Fig. 7. Hormone induction of ADF9p::GUS expression and adf9-1 defective callus formation.
(a) ADF9p::GUS is a reporter for ADF9 promoter and intron localized enhancer activity (Ruzicka et al. 2007). (b–g) 12-day-old seedlings containing the ADF9p::GUS promoter-reporter fusion were subjected to 24 h treatment with phytohormones indicated, assayed briefly for β-glucuronidase (GUS) activity, and compared to seedlings treated only with DMSO, the solvent for each hormone. (b) DMSO control (~ 1 mM). (c) 10 µM 2,4,-dichlorophenoxyacetic acid (2,4,-D). (d) 10 µM gibberellic acid (GA); (e) 10 µM abscisic acid (ABA); (f) 10 µM kinetin; (g) 10 µM indolacetic acid (IAA). (h and i). Samples of callus formed from root tissue are compared between wild-type (h) and adf9-1 (i). See Fig. 3 for quantification of this phenotype
Callus induction from ADF9-defective tissue
Mutants in the phytohormone-induced actin ACT7 are defective in callus formation (Kandasamy et al. 2001). We considered the possibility that if phytohormones control ADF9 and its role in organ development, ADF9-defective plants might also exhibit growth defects when organ explants were grown on callus-inducing media. To test this proposal, identically sized root sections were removed from vertically grown wild-type and adf9-1 mutant plants and placed on media containing 2,4-dichlorophenoxyacetic acid (2,4-D) and kinetin. The untreated mutant and wild-type roots were indistinguishable in morphology when examined under a dissecting microscope. After 4 weeks, the wild-type callus tissue consisted of large clumps of cells, illustrated in Fig. 7h, comprised of multiple cell types (not shown). The adf9-1 mutant exhibited a significant difference in callus morphology compared to wild-type, with fewer aggregates of large cells and more and longer root hair-like cells (Fig. 7i). Additionally, the average fresh weight of the adf9-1 callus samples was less than half of wild-type callus samples (Fig. 3d). The A2pt::ADF9 cDNA expression construct restored the wild-type callus growth rate (Fig. 3d) and morphology to the adf9-1 mutant (not shown).
Discussion
Distinct functions for ADF9 in the shoot apical meristem
Defects in organ initiation, growth rates, and flowering time in the ADF9 mutants are all consistent with a disruption of activities in the SAM (Carraro et al. 2006; Guyomarc’h et al. 2005). Among the 11 diverse members of the Arabidopsis ADF family, there is considerable potential for functional redundancy in the SAM. The ADF5 and ADF9 genes comprise class III ADFs. The ADF9 isovariant differs from ADF5 in 29 of its 139 amino acids, representing considerable divergence for proteins in the same class (Ruzicka et al. 2007). While both genes are expressed in the SAM, each has its own distinct expression phenotype. ADF9 is moderately expressed in the SAM and induced by several phytohormones, while ADF5 is not hormone responsive and is expressed more broadly in vegetative tissues. In addition to ADF9 and ADF5, four vegetative class I ADF genes (ADF1, 2, 3, 4) and the constitutive class IV ADF6 are expressed in the SAM, all more strongly than ADF9 (Ruzicka et al. 2007). The expression of seven ADFs from three divergent classes in the SAM suggests there must be some redundancy among their activities. However, the apparent SAM-associated ADF9-deficient phenotypes and ADF9’s divergent protein sequence argue that this isovariant is functionally distinct from the other ADF isovariants expressed in the SAM. We have confirmed that distant ADF4 and ADF8 cDNAs expressed from the constitutive A2pt cassette do not complement the adf9-1 phenotypes (data not shown).
The F-actin cytoskeleton was somewhat abnormal in trichomes of the adf9-1 mutant (not shown) and is most likely abnormal in other cell types where ADF9 is expressed. Considering the roles that the ADF-controlled actin cytoskeleton is expected to play in elaborating pathways of growth and differentiation, loss-of-function mutations in one or more ADFs expressed in the SAM would be expected to cause developmental defects (Jurgens 2005). The actin cytoskeleton is essential to the construction of the preprophase band that positions the division plane and determines cell polarity (Gallagher and Smith 2000; Smith 2001). Cell polarity is arguably one of the most important factors in organ initiation and growth. Each of the phenotypes observed for the adf9-1 allele might be attributed to changes in the cytoplasmic actin cytoskeleton within critical layers of cells in the SAM that determine leaf and stem growth rates and the initiation of lateral branches and the inflorescence (Carraro et al. 2006; Castellano and Sablowski 2005). Organ initiation and outgrowth from the SAM may be modeled as occurring in three stages: stage (1) signaling among cells of the meristem; stage (2) development of primordial cells; and stage (3) establishment of cell polarity and organ outgrowth. Potential ADF9-related defects in the cytoskeleton of SAM cells might be expected to alter the latter two stages. Furthermore, auxin, cytokinin, and GA all affect organ development from the SAM (Carraro et al. 2006; Castellano and Sablowski 2005), perhaps linking the observed hormone-induced expression of ADF9 with the loss of apical dominance and fewer and smaller leaves at the time of flowering in ADF9 defective plants. In addition, these hormones are known to stimulate cytoskeletal dynamics and membrane assembly both in the formation of the cell plate required for cytokinesis and in subsequent cell expansion, leading to outgrowth of new organs (stages 2 & 3) (Jurgens 2005). For example, GA stimulates activators of the transition to flowering and this pathway may ultimately stimulate growth via activities in the cytoskeleton. Thus, the stimulation of ADF9 expression by hormones may be essential to restructuring the cytoskeleton in appropriate meristem cells and other tissues during organ initiation.
Nuclear vs cytoplasmic ADF activities controlling gene expression
ADF/cofilins have the potential to alter nuclear gene expression and down stream multicellular development via their activities in either the nucleus or cytoplasm. First, consider the activities identified for conventional actin and ADF in the nucleus (Bettinger et al. 2004; Gettemans et al. 2005; Miralles and Visa 2006; Visa 2005). Nuclear actin is a stoichiometric component of some chromatin remodeling complexes isolated from yeast and humans (Mizuguchi et al. 2004; Olave et al. 2002; Ruhl et al. 2006). In addition, actin participates in transcriptional initiation via interactions with RNA polymerase (Pol) I, II, and III, is localized to Cajal bodies with a likely role in RNA processing, and functions in the export of ribonuclear particles (RNPs) and mRNAs. Mammalian and higher plant ADFs contain a conserved nuclear localization motif and several have been localized to the nucleus (Bamburg 1999; Ohta et al. 1989; Ruzicka et al. 2007). In mammalian cells, the nuclear import of ADF appears to be essential to the nuclear import of actin (Abe et al. 1993; Pendleton et al. 2003). For example, ADF antibodies microinjected into the cytoplasm block the nuclear import of actin. The phosphorylated form of ADF is thought to be inactive in cytoplasmic activities, but is preferentially localized to the nucleus (Nebl et al. 1996). The phosphorylation state of ADF is highly regulated by specific kinases and phosphatases, and hence, the nuclear import of ADF is highly regulated, perhaps to control the import of actin or other factors and network their role in gene expression (Sarmiere and Bamburg 2004; Tanaka et al. 2005a; Tanaka et al. 2005b). No direct role for ADF in nuclear control of gene expression has yet been reported (i.e., acting as a transcription factor or component of a chromatin remodeling complex). We have not detected significant levels of nuclear actin in Arabidopsis cells using a variety of plant actin-specific monoclonal antibodies (unpublished observations), however actin conformation-specific antibodies have detected actin in isolated plant nuclei (Cruz et al. 2008).
In addition to possible nuclear activities, ADF/cofilins have the potential to control gene expression by their activities in the cytoplasm. A general model is emerging in which activities that stimulate F-actin polymerization or depolymerization release cytoplasmic actin-bound nuclear cofactors. For example, human Cofilin 1 functions as a repressive cofactor of glucocorticoid receptor (GR). Apparently, through cofilin’s ability to promote actin turnover, cofilin releases cytoplasmic GR bound to an actin-hsp90 complex, promotes the redistribution of GR to the nucleus, and causes the activation of GR target gene expression (Ruegg et al. 2004). Furthermore, activities that stimulate F-actin polymerization release other cytoplasmic G-actin-bound transcriptional cofactors, as reported for the co-activators of serum response factor SRF (Vartiainen et al. 2007). In addition, we have shown that Arabidopsis HEXOKINASE1 binds F-actin and its nuclear signalling is disrupted by activities that depolymerize F-actin (Balasubramanian et al. 2007). In summary, the ADF/cofilins have the potential to alter multicellular development via activities in the nucleus or the cytoplasm that control gene expression. In the case of ADF9, this might alter signaling among of cells in the SAM and alter organ initiation (stage I). These activities would be distinct from cytoskeletal roles for ADF9 in later stages of SAM development. Although these previous published plant and animal studies indicating roles for ADFs in controlling transcription factor distribution and target gene expression have been carried out at the cellular level, they suggest models as to how ADF9 might regulate early stages of multicellular development.
The role of Arabidopsis ADF9 in development should also be considered in light of the mounting evidence that critical steps in leaf, shoot, and floral organ initiation from the SAM are controlled at the level of chromatin remodeling (Guyomarc’h et al. 2005; He and Amasino 2005; Meagher et al. 2007; Williams and Fletcher 2005). ADF9 is most strongly expressed in the SAM (Ruzicka et al. 2007). The SAM gives rise not only to leaves, but to inflorescence meristems and to floral meristems, and critical steps in each developmental phase are regulated at the level of chromatin structure. The seedling, lateral branch, and flowering time phenotypes of ADF9-defective plants could be ascribed to chromatin remodeling defects that affect the earliest stage of organ development, when morphogenic information is exchanged among cells within the SAM (stage I). For particular examples, defects in homologs of well-characterized chromatin remodeling components Arabidopsis ACTIN-RELATED PROTEIN4 and 6, PHOTOPERIOD INDEPENDENT1, SERRATED LEAVES AND EARLY FLOWERING and EARLY FLOWERING7 and 8 each produce early flowering time phenotypes (Choi et al. 2007; He et al. 2004; March-Diaz et al. 2007; Meagher et al. 2007).
We were assisted in our exploration of the role of ADF9 in chromatin-level control of early flowering, because there are robust data demonstrating epigenetic controls over the activity of the FLC locus and FLC’s role in the repression of the transition to flowering. We made four measurements of chromatin structure at FLC. Using ChIP assays we showed decreased acetylation and trimethylation of lysines in histone H3 at most of the nucleosomal regions assayed and decreased H2A.Z deposition within nucleosomes in the promoter region. Using a nucleosome scanning assay for nucleosomal density we identified a 300 bp nucleosome depleted region within the wild-type FLC promoter located immediately upstream of the transcriptional start site. Mononucleosome enriched preparations from the adf9-1 mutant showed significantly decreased nuclease sensitivity across this region of the FLC locus, but not in flanking sequences, relative to wild-type. Our data are consistent with a several fold increase in nucleosome density in this region in adf9-1. All four assays suggest that defects in ADF9 expression cause the FLC locus to take on a less permissive chromatin structure. The precise mechanisms and machinery through which ADF9 contributes to these changes in chromatin structure and whether ADF9 is acting from the nucleus or cytoplasm will be the subject of future studies.
Dissection of the flowering time signaling pathway
Based on the early flowering time phenotypes and the expression of FLC, FT, SOC, and LFY transcripts in mutant and/or complemented plant lines, it is logical to model ADF9 as an indirect activator of FLC, and hence, a repressor of flowering time (Fig. 4a) (He and Amasino 2005). For example, via its role transporting actin or other factors into the nucleus, ADF9 might be essential to the activity of chromatin remodeling complexes that activate FLC expression. Consistent with this indirect role for ADF9, the overexpression of ADF9 cDNA in the adf9-1 complemented lines did not result in the overexpression of FLC or late flowering. Note that the overexpression of FLC is known to dramatically delay flowering (Michaels and Amasino 1999). Chromatin remodeling complexes are typically composed of 10 or more subunits (Bao and Shen 2007; Olave et al. 2002). Thus, if ADF9 increased the concentration of one or a few factors in the nucleus it is unlikely to alter the activities of such complex machines.
Some aspects of the ADF9-deficient early flowering phenotype cannot be reconciled with current knowledge about the flowering time signaling pathway. In particular, the photoperiod-dependent flowering pathway in Arabidopsis is thought to function independent of FLC levels, typically through CO and its downstream targets, FT and SOC1 (Fig. 4a) (Valverde et al. 2004; Yanovsky and Kay 2002). CO transcripts were modestly induced in adf9-1, and more interestingly CO levels and one of its targets, FT, were significantly down-regulated below wild-type levels, when the mutant was complemented with ADF9 expression from the A2pt:ADF9 transgene. However, we have not shown that changes in CO-levels account for the photoperiod-dependent early flowering phenotype. Thus, it appears that ADF9 acts at some level as a novel repressor of photoperiod-dependent early flowering by networking regulation through both CO and FLC and perhaps other unknown factors. Future genetic and biochemical studies will attempt to unravel this distinct pattern of altered gene expression.
Conclusion
ADF9 encodes distinct protein isovariant within the diverse Arabidopsis ADF gene family. ADF9 deficiencies produced morphological and cytoskeletal defects in growth, branching, and F-actin filament structure consistent with ADF’s role in remodeling the actin cytoskeleton in cells in the SAM affecting cytokinesis and cell and organ expansion. However, we also demonstrated that ADF9 deficiency produced early flowering, gene expression, and chromatin remodeling phenotypes consistent with defects in earliest stages of SAM development. Our data suggest more generally that plant ADFs and the associated actin cytoskeleton might network cytoplasmic and nuclear activities that control multicellular development.
Supplementary Material
Acknowledgements
We thank Dr. M. Kandasamy and Gay Gragson for critical reading and discussions of the manuscript and Dr. Richard Amasino for helpful discussions on flowering time experiments. This work was supported by funding from the National Institutes of Health (5RO1GM036397-22) to R.B.M.; an NIH supplement (3RO1GM036397-20S1/C103JD) to B.B; and NIH training grant (GM 07103-29) support to D.R.R., R.B.D., and L.K.-R.
Footnotes
Richard B. Meagher is responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors.
Brunilís Burgos-Rivera and Daniel R. Ruzicka made equivalent contributions to the manuscript.
Electronic supplementary material The online version of this article (doi:10.1007/s11103-008-9398-1) contains supplementary material, which is available to authorized users.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Brunilís Burgos-Rivera, Department of Genetics, University of Georgia, Athens, GA 30602, USA.
Daniel R. Ruzicka, Donald Danforth Plant Science Centre, St. Louis, MO 63132, USA
Roger B. Deal, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave North, Seattle, WA 98109, USA
Elizabeth C. McKinney, Department of Genetics, University of Georgia, Athens, GA 30602, USA
Lori King-Reid, Department of Genetics, University of Georgia, Athens, GA 30602, USA.
Richard B. Meagher, Email: meagher@uga.edu, Department of Genetics, University of Georgia, Athens, GA 30602, USA
References
- Abe H, Nagaoka R, Obinata T. Cytoplasmic localization and nuclear transport of cofilin in cultured myotubes. Exp Cell Res. 1993;206:1–10. doi: 10.1006/excr.1993.1113. doi: 10.1006/excr.1993.1113. [DOI] [PubMed] [Google Scholar]
- Balasubramanian R, Karve A, Kandasamy M, Meagher RB, Moore BD. A role for F-actin in hexokinase-mediated glucose signaling. Plant Physiol. 2007;145:1423–1434. doi: 10.1104/pp.107.108704. doi: 10.1104/pp.107.108704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamburg JR. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- Bao Y, Shen X. SnapShot: chromatin remodeling complexes. Cell. 2007;129:632. doi: 10.1016/j.cell.2007.04.018. doi: 10.1016/j.cell.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- Bettinger BT, Gilbert DM, Amberg DC. Actin up in the nucleus. Nat Rev Mol Cell Biol. 2004;5:410–415. doi: 10.1038/nrm1370. doi: 10.1038/nrm1370. [DOI] [PubMed] [Google Scholar]
- Carraro N, Peaucelle A, Laufs P, Traas J. Cell differentiation and organ initiation at the shoot apical meristem. Plant Mol Biol. 2006;60:811–826. doi: 10.1007/s11103-005-2761-6. doi: 10.1007/s11103-005-2761-6. [DOI] [PubMed] [Google Scholar]
- Castellano MM, Sablowski R. Intercellular signalling in the transition from stem cells to organogenesis in meristems. Curr Opin Plant Biol. 2005;8:26–31. doi: 10.1016/j.pbi.2004.11.010. doi: 10.1016/j.pbi.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Choi K, Park C, Lee J, Oh M, Noh B, Lee I. Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development. 2007;134:1931–1941. doi: 10.1242/dev.001891. doi: 10.1242/dev.001891. [DOI] [PubMed] [Google Scholar]
- Cruz JR, de la Torre C, Moreno Diaz, de la Espina S. Nuclear actin in plants. Cell Biol Int. 2008;32:584–587. doi: 10.1016/j.cellbi.2007.11.004. doi: 10.1016/j.cellbi.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Deal RB, Topp CN, McKinney EC, Meagher RB. Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A. Z. Plant Cell. 2007;19:74–83. doi: 10.1105/tpc.106.048447. doi: 10.1105/tpc.106.048447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL, Brown AHD, Grace JP. Multiple origins of polyploids in the Glycine tabacina complex inferred from chloroplast DNA polymorphism. Proc Natl Acad Sci USA. 1990;87:714–717. doi: 10.1073/pnas.87.2.714. doi: 10.1073/pnas.87.2.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher K, Smith LG. Roles for polarity and nuclear determinants in specifying daughter cell fates after an asymmetric cell division in the maize leaf. Curr Biol. 2000;10:1229–1232. doi: 10.1016/s0960-9822(00)00730-2. doi: 10.1016/S0960-9822(00)00730-2. [DOI] [PubMed] [Google Scholar]
- Gaudin V, Libault M, Pouteau S, Juul T, Zhao G, Lefebvre D, et al. Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development. 2001;128:4847–4858. doi: 10.1242/dev.128.23.4847. [DOI] [PubMed] [Google Scholar]
- Gettemans J, Van Impe K, Delanote V, Hubert T, Vandekerckhove J, De Corte V. Nuclear actin-binding proteins as modulators of gene transcription. Traffic. 2005;6:847–857. doi: 10.1111/j.1600-0854.2005.00326.x. doi: 10.1111/j.1600-0854.2005.00326.x. [DOI] [PubMed] [Google Scholar]
- Gilliland LU, McKinney EC, Asmussen MA, Meagher RB. Detection of deleterious genotypes in multigenerational studies. I. Disruptions in individual Arabidopsis actin genes. Genetics. 1998;149:717–725. doi: 10.1093/genetics/149.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyomarc’h S, Bertrand C, Delarue M, Zhou DX. Regulation of meristem activity by chromatin remodelling. Trends Plant Sci. 2005;10:332–338. doi: 10.1016/j.tplants.2005.05.003. doi: 10.1016/j.tplants.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. From genes to plants via meristems. Development. 2005;132:2679–2684. doi: 10.1242/dev.01880. doi: 10.1242/dev.01880. [DOI] [PubMed] [Google Scholar]
- He Y, Amasino RM. Role of chromatin modification in flowering-time control. Trends Plant Sci. 2005;10:30–35. doi: 10.1016/j.tplants.2004.11.003. doi: 10.1016/j.tplants.2004.11.003. [DOI] [PubMed] [Google Scholar]
- He Y, Doyle MR, Amasino RM. PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 2004;18:2774–2784. doi: 10.1101/gad.1244504. doi: 10.1101/gad.1244504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto A, Baum B. Actin in development. Mech Dev. 2003;120:1337–1349. doi: 10.1016/j.mod.2003.06.006. doi: 10.1016/j.mod.2003.06.006. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens G. Cytokinesis in higher plants. Annu Rev Plant Biol. 2005;56:281–299. doi: 10.1146/annurev.arplant.55.031903.141636. doi: 10.1146/annurev.arplant.55.031903.141636. [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, Gilliland LU, McKinney EC, Meagher RB. One plant actin isovariant, ACT7, is induced by auxin and required for normal callus formation. Plant Cell. 2001;13:1541–1554. doi: 10.1105/TPC.010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Balish RS, Heaton AC, McKinney EC, Dhankher OP, Meagher RB. Engineering a root-specific, repressor-operator gene complex. Plant Biotechnol J. 2005;3:571–582. doi: 10.1111/j.1467-7652.2005.00147.x. doi: 10.1111/j.1467-7652.2005.00147.x. [DOI] [PubMed] [Google Scholar]
- Li Y, Dhankher O, Carreira L, Balish R, Meagher R. Arsenic and mercury tolerance and cadmium sensitivity in plants expression of bacterial gamma-glutamylcysteine synthetase. Environ Toxicol Chem. 2005;24:1376–1386. doi: 10.1897/04-340r.1. doi: 10.1897/04-340R.1. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maciver SK, Hussey PJ. The ADF/cofilin family: actin-remodeling proteins. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-5-reviews3007. reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March-Diaz R, Garcia-Dominguez M, Florencio FJ, Reyes JC. SEF, a new protein required for flowering repression in Arabidopsis, interacts with PIE1 and ARP6. Plant Physiol. 2007;143:893–901. doi: 10.1104/pp.106.092270. doi: 10.1104/pp.106.092270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Trillo M, Lazaro A, Poethig RS, Gomez-Mena C, Pineiro MA, Martinez-Zapater JM, et al. EARLY IN SHORT DAYS 1 (ESD1) encodes ACTIN-RELATED PROTEIN 6 (AtARP6), a putative component of chromatin remodelling complexes that positively regulates FLC accumulation in Arabidopsis. Development. 2006;133:1241–1252. doi: 10.1242/dev.02301. doi: 10.1242/dev.02301. [DOI] [PubMed] [Google Scholar]
- Mathur J. Cell shape development in plants. Trends Plant Sci. 2004;9:583–590. doi: 10.1016/j.tplants.2004.10.006. doi: 10.1016/j.tplants.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Meagher RB, Fechheimer M. The cytoskeletal proteome of Arabidopsis. In: Meyerowitz E, Somerville C, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- Meagher RB, Kandasamy MK, Deal RB, McKinney EC. Actin-related proteins in chromatin-level control of the cell cycle and developmental transitions. Trends Cell Biol. 2007;17:325–332. doi: 10.1016/j.tcb.2007.06.001. doi: 10.1016/j.tcb.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakhina S, Myers R, Druzhinina M, Steward R. Crosstalk between the actin cytoskeleton and Ran-mediated nuclear transport. BMC Cell Biol. 2005;6:32. doi: 10.1186/1471-2121-6-32. doi: 10.1186/1471-2121-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F, Visa N. Actin in transcription and transcription regulation. Curr Opin Cell Biol. 2006;18:261–266. doi: 10.1016/j.ceb.2006.04.009. doi: 10.1016/j.ceb.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Plant Physiol. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [Google Scholar]
- Nebl G, Meuer SC, Samstag Y. Dephosphorylation of serine 3 regulates nuclear translocation of cofilin. J Biol Chem. 1996;271:26276–26280. doi: 10.1074/jbc.271.42.26276. doi: 10.1074/jbc.271.42.26276. [DOI] [PubMed] [Google Scholar]
- Ohta Y, Nishida E, Sakai H, Miyamoto E. Dephosphorylation of cofilin accompanies heat shock-induced nuclear accumulation of cofilin. J Biol Chem. 1989;264:16143–16148. [PubMed] [Google Scholar]
- Olave IA, Reck-Peterson SL, Crabtree GR. Nuclear actin and actin-related proteins in chromatin remodeling. Annu Rev Biochem. 2002;71:755–781. doi: 10.1146/annurev.biochem.71.110601.135507. doi: 10.1146/annurev.biochem.71.110601.135507. [DOI] [PubMed] [Google Scholar]
- Pawloski LC, Kandasamy MK, Meagher RB. The late pollen actins are essential for normal male and female development in Arabidopsis. Plant Mol Biol. 2006;62:881–896. doi: 10.1007/s11103-006-9063-5. doi: 10.1007/s11103-006-9063-5. [DOI] [PubMed] [Google Scholar]
- Pendleton A, Pope B, Weeds A, Koffer A. Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells. J Biol Chem. 2003;278:14394–14400. doi: 10.1074/jbc.M206393200. doi: 10.1074/jbc.M206393200. [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- Ruegg J, Holsboer F, Turck C, Rein T. Cofilin 1 is revealed as an inhibitor of glucocorticoid receptor by analysis of hormone-resistant cells. Mol Cell Biol. 2004;24:9371–9382. doi: 10.1128/MCB.24.21.9371-9382.2004. doi: 10.1128/MCB.24.21.9371-9382.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl DD, Jin J, Cai Y, Swanson S, Florens L, Washburn MP, et al. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry. 2006;45:5671–5677. doi: 10.1021/bi060043d. doi: 10.1021/bi060043d. [DOI] [PubMed] [Google Scholar]
- Ruzicka DR, Kandasamy MK, McKinney EC, Burgos-Rivera B, Meagher RB. The ancient subclasses of Arabidopsis ACTIN DEPOLYMERIZING FACTOR genes exhibit novel and differential expression. Plant J. 2007;52:460–472. doi: 10.1111/j.1365-313X.2007.03257.x. doi: 10.1111/j.1365-313X.2007.03257.x. [DOI] [PubMed] [Google Scholar]
- Sarmiere PD, Bamburg JR. Regulation of the neuronal actin cytoskeleton by ADF/cofilin. J Neurobiol. 2004;58:103–117. doi: 10.1002/neu.10267. doi: 10.1002/neu.10267. [DOI] [PubMed] [Google Scholar]
- Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Smith LG. Plant cell division: building walls in the right places. Nat Rev Mol Cell Biol. 2001;2:33–39. doi: 10.1038/35048050. doi: 10.1038/35048050. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Nishio R, Haneda K, Abe H. Functional involvement of Xenopus homologue of ADF/cofilin phosphatase, slingshot (XSSH), in the gastrulation movement. Zool Sci. 2005a;22:955–969. doi: 10.2108/zsj.22.955. doi: 10.2108/zsj.22.955. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Okubo Y, Abe H. Involvement of slingshot in the Rho-mediated dephosphorylation of ADF/cofilin during Xenopus cleavage. Zool Sci. 2005b;22:971–984. doi: 10.2108/zsj.22.971. doi: 10.2108/zsj.22.971. [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–1752. doi: 10.1126/science.1141084. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- Vega-Palas MA, Ferl RJ. The Arabidopsis Adh gene exhibits diverse nucleosome arrangements within a small DNase I-sensitive domain. Plant Cell. 1995;7:1923–1932. doi: 10.1105/tpc.7.11.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit B. Stem cell signalling networks in plants. Plant Mol Biol. 2006;60:793–810. doi: 10.1007/s11103-006-0033-8. doi: 10.1007/s11103-006-0033-8. [DOI] [PubMed] [Google Scholar]
- Visa N. Actin in transcription. Actin is required for transcription by all three RNA polymerases in the eukaryotic cell nucleus. EMBO Rep. 2005;6:218–219. doi: 10.1038/sj.embor.7400362. doi: 10.1038/sj.embor.7400362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YS, Motes CM, Mohamalawari DR, Blancaflor EB. Green fluorescent protein fusions to Arabidopsis fimbrin 1 for spatio-temporal imaging of F-actin dynamics in roots. Cell Motil Cytoskeleton. 2004;59:79–93. doi: 10.1002/cm.20024. doi: 10.1002/cm.20024. [DOI] [PubMed] [Google Scholar]
- Williams L, Fletcher JC. Stem cell regulation in the Arabidopsis shoot apical meristem. Curr Opin Plant Biol. 2005;8:582–586. doi: 10.1016/j.pbi.2005.09.010. doi: 10.1016/j.pbi.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA. Molecular basis of seasonal time measurement in Arabidopsis. Nature. 2002;419:308–312. doi: 10.1038/nature00996. doi: 10.1038/nature00996. [DOI] [PubMed] [Google Scholar]
- GENEVESTIGATOR. Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. Arabidopsis microarray database and analysis toolbox ( www.genevestigator.ethz.ch) Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.