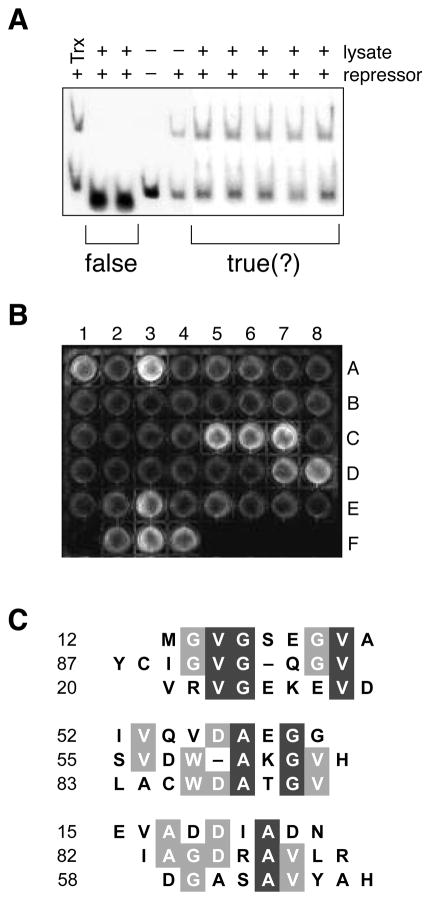
Figure 2.
In vitro screens to identify true positives from the genetic selection. A, Representative gel retardation assay. False positives that interfere with the protein–DNA interaction were identified by using a gel retardation assay to monitor the binding of cI to λOR1 in the presence of Trx–peptide fusions. B, Representative high-throughput ELISA. Trx–peptide fusions not identified as false positives by the gel retardation assay were screened for binding to immobilized monomeric protease. A1 through E8 are protease-coated surfaces to which 40 lysates were applied. Binding was detected with an anti-Trx antibody. F1 is a surface coated with free cysteine to which no lysate was applied. F2 is a protease-coated surface to which a lysate containing Trx with no fused peptide was applied. F3 is a protease-coated surface to which no lysate was applied and anti-protease antibody (S. Oroszlan) was used instead of anti-Trx antibody. F4 is a Trx-coated surface to which no lysate was applied. C, Sequence alignment of positive isolates. The 9 sequences that gave the strongest signal in the ELISA were divided into 3 subgroups according to their similarity. Residues that are conserved partially are in gray boxes; those conserved completely are in black boxes. The alignment was made with the MULTIALIGN interface of the BLOSOM62 algorithm55.