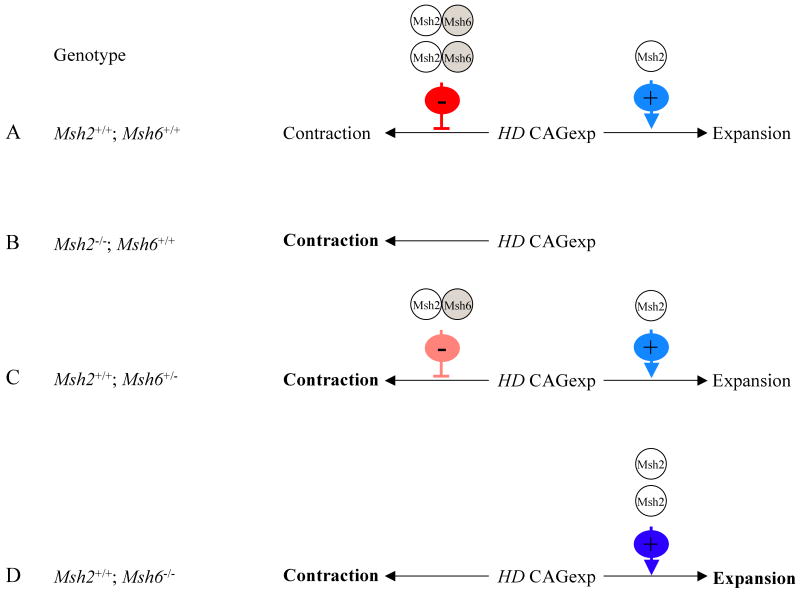
Figure 3.
Nuclear mutant huntingtin immunostaining in striatal neurons.
HdhQ111 mice were crossed onto backgrounds deficient in Msh2 (A), Xpc (B), Msh6 (C) or Msh3 (D). Striatal sections from HdhQ111/+ mice at 5 months of age were immunostained with anti-huntingtin antibody EM48 and a staining index (SI) calculated for each mouse as the product of the mean staining intensity and the number of immunostained nuclei (see Materials and Methods).
Left: Representative striatal histological sections immunostained with EM48.
Right: Bar graphs showing the mean SI for each genotype +/- standard error.
The number of mice of each DNA repair genotype (+/+, +/-, -/-) and the constitutive HD CAG repeat size determined from tail in each mouse is as follows: (A) Msh2+/+ n=3, CAG 107, 108, 109; Msh2+/- n=4, CAG 106, 106, 107, 107. (B) Xpc+/+ n=4, CAG 99, 101, 101, 101; Xpc-/- n=3, CAG 98, 101, 102. (C) Msh6+/+ n=3, CAG 98, 99, ND#; Msh6+/- n=4, CAG 97, 98, 99, 99; Msh6-/- n=4, CAG 98, 99, 99, 99. (D) Msh3+/+ n=5, CAG 98, 100, 100, 101, 102; Msh3+/- n=5, CAG 97, 97, 100, 101, 105; Msh3-/- n=3, CAG 99, 100, 102. * p=0.02, ** p<0.001. #A repeat value could not be determined for this mouse. Loss of two Msh3 alleles had a slightly greater effect of loss of one Msh3 allele but this effect was not statistically significant (p=0.11).