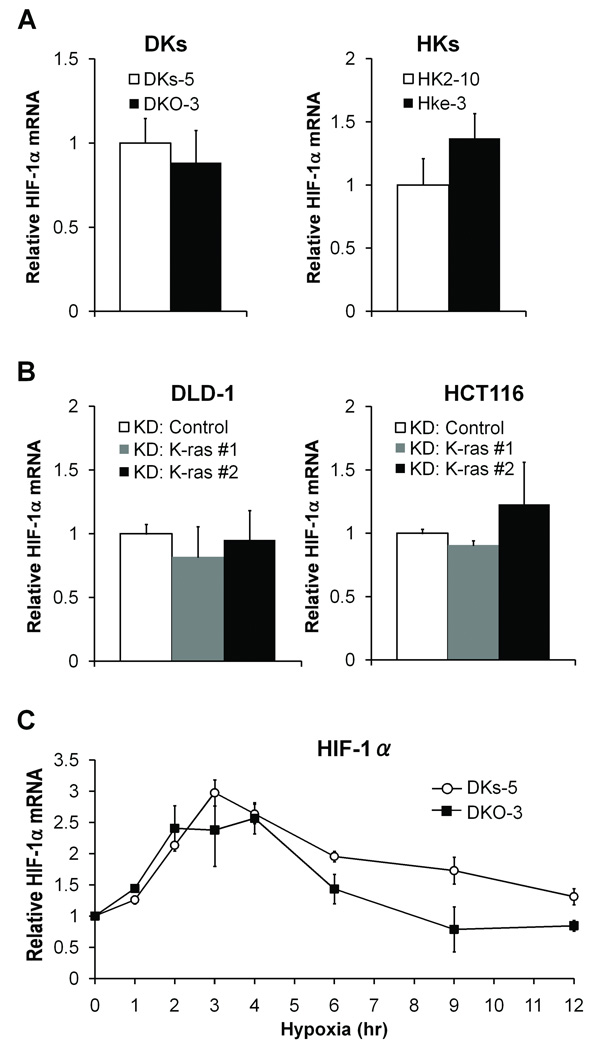
Figure 2.
Depletion of mutant KRAS and BRAF oncogenes and hypoxic induction of HIF proteins. Hypoxic induction of HIF-1α and HIF-2α proteins was analyzed by western blotting. A, DKs-5, DKO-3, HK2-10 and HKe-3 cells were incubated in either normoxia (N) or hypoxia (H) for 12 hours. The weak K-ras bands seen in the DKO-3 and Hke-3 cells correspond to the remaining wild-type KRAS allele. B, DLD-1 and HCT116 cells were treated with siRNA targeting K-ras (K-ras #1 and #2) or control siRNA. Thirty-six hours after transfection, cells were subjected to either normoxia (N) or hypoxia (H) for 12 hours. C, shRNA against BRAF (shBRAF #1 and #2) or empty vector was lentivirally transduced into HT29 cells. Stable cells (HT29 pSicoR) were incubated in either normoxia (N) or hypoxia (H). D, HT29 cells were treated with siRNA targeting BRAF (BRAF #1 and #2) or control siRNA, and then cells were subjected to either normoxia (N) or hypoxia (H). Band densities were quantified and HIF protein levels were normalized to β-actin. Numbers below the bottom panel represent the mean fold change in hypoxia relative to control for 2 independent experiments. *P<0.05.