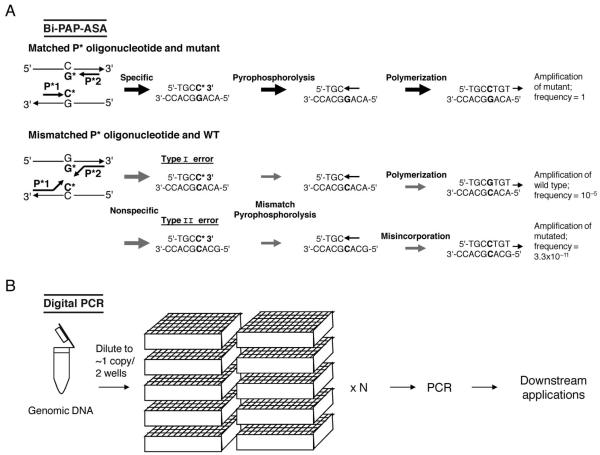
Fig. 1. Highly selective PCR-based methods for known and unknown mutation enhancement and identification.
(A), Bidirectional pyrophosphorolysis-activated polymerization allele-specific amplification (bi-PAP-ASA) for high selectivity of known mutations (26). P* is a specifically designed oligonucleotide with a 3′-terminal blocker that is activated, but not extended, by pyrophosphorolysis. Downstream and upstream P* contain dideoxy C and G at the 3′ termini that are specific to the mutant but not the wild type. Efficient amplification of the mutant occurs after pyrophosphorolysis (to remove the 3′-terminal ddCMP) and polymerization. Inefficient amplification is denoted by the gray arrows. Nonspecific type I error amplification is rare; type II error is caused by serial mismatch phosphorolysis and misincorporation, which results in the exponential amplification of the mutated product and reduces selectivity. (B), Digital PCR for high selectivity of both known and unknown mutations (27). Genomic DNA is diluted to approximately 1–2 copies per well. The number (N) of required wells varies widely and depends on putative mutant and wild-type concentrations. PCR is performed on each sample well individually. PCR amplicons can be used in many downstream applications such as direct sequencing, pyrosequencing, TaqMan assays, and molecular beacons.