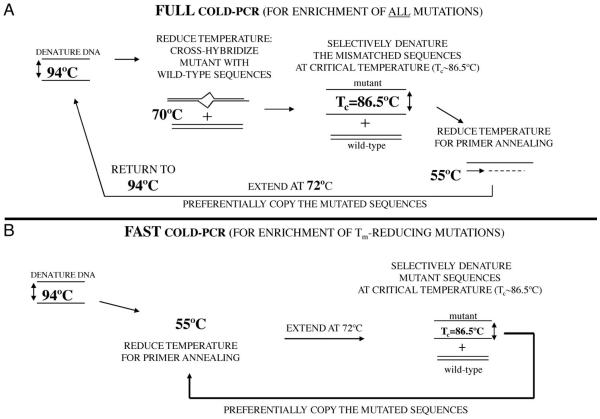
Fig. 2. COLD-PCR protocol (40). Two forms of COLD-PCR have been developed, Full COLD-PCR (A) and Fast COLD-PCR (B). An example protocol for a 167-bp TP53 amplicon is reviewed here.
(A), Full COLD-PCR has the potential to enrich all possible mutations. Several preliminary rounds of conventional PCR enable an initial buildup of 1 or more target amplicons, then the cycling switches to COLD-PCR. After denaturation at 94 °C, the PCR amplicons are incubated (e.g., 70 °C for 2–8 min) for reannealing and cross-hybridization. Cross-hybridization of mutant and wild-type alleles forms a mismatch-containing structure (heteroduplex) that has a lower melting temperature than a fully-matched structure (homoduplex). The PCR temperature is next raised to the critical denaturation temperature (Tc) (e.g., 86.5 °C) to preferentially denature the heteroduplexed amplicons. The temperature is reduced for primer annealing (e.g., 55 °C) and then raised to 72 °C to extend the amplicon and preferentially amplify the mutation-containing alleles. (B), Fast COLD-PCR is a simpler cycling that can be performed to enrich for mutations that reduce the melting temperature of the wild-type amplicon. Using the mutant Tc, rather than the standard 94 °C denaturation temperature, preferentially denatures the lower-Tm allele. Fast COLD-PCR does not perform the 70 °C incubation step. Fast COLD-PCR amplification and enrichment begins earlier in the cycling than in full COLD-PCR, thus resulting in higher enrichment.