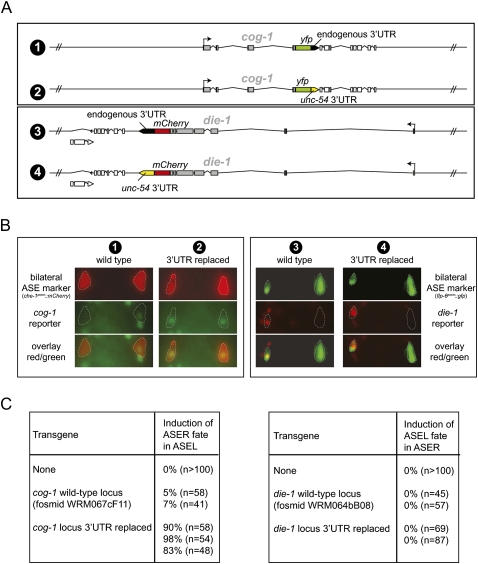
FIGURE 8.
Differential contribution of the 3′UTR for regulation of expression and activity of the die-1 and cog-1 gene. (A) Schematic representation of the fosmid clones used for expression (panel B) and functional (panel C) analysis of die-1 and cog-1. Fosmid clones are around about 40 kbp in size. Fluorescent reporters were first recombineered into the respective fosmids. In a subsequent recombineering step, 3′UTRs were replaced with the unc-54 3′UTR as schematically indicated. Fosmids were all injected at similar concentration to generate transgenic animals. (B) Testing the effect of 3′UTR replacement on the left/right asymmetric expression of die-1 and cog-1. Representative yfp/mCherry expression of transgenic animals expressing the reporter genes shown in panel A. At least two independent transgenic lines were tested for expression, and expression was found to be almost fully penetrant in that, for the cog-1 reporters, the construct with wild-type 3′UTR was asymmetric while the reporter with the unc-54 3′UTRs was not. For the die-1 reporters, both always showed left/right asymmetric expression regardless of the 3′UTR tested. Bilateral markers (flp-6prom∷cameleon = ntIs13 and che-1prom∷mCherry = otIs232) were used to unambiguously identify the ASE neurons (highlighted with stippled lines). Numbers above each panel correspond to the constructs shown in panel A. (C) Testing the effect of 3′UTR replacement on functional derepression of die-1 and cog-1 gene activity. Based on previous work, derepression of die-1 function in ASER is expected to result in a transformation of ASER to ASEL (Johnston et al. 2005), while derepression of cog-1 function in ASEL is expected to result in a transformation of ASEL to ASER (Chang et al. 2003; Johnston et al. 2005; Sarin et al. 2009). Transgenic animals containing reporters with wild-type 3′UTRs or replaced 3′UTR (as shown in panel A) were assayed for their ability to convert cell fate, as assessed with the ASER fate marker gcy-5∷gfp (ntIs1), which is expressed in ASEL if ASER fate is induced (left table) and repressed in ASER if ASEL fate is induced (right table). Each row represents an independent transgenic line. For die-1, we also performed the same experiment with fosmids in which the locus was not tagged with a fluorescent reporter. In that case zero of four lines with the wild-type 3′UTR induced ASEL fate in ASER, and one of four lines with the replaced 3′UTR induced ASEL in ASER in 25% (n = 89) of animals.