Summary
Low folate status may be a consequence of suboptimal intake, transport or cellular utilization of folate and, together with elevated homocysteine, is a recognized risk factor/marker for several human pathologies. As folate transport across cell membranes is mediated in part by the reduced folate carrier (RFC1), variants within this gene may influence disease risk via an effect on folate and/or homocysteine levels. The present study was undertaken to assess the association between the SLC19A1 (RFC1) c.80G>A polymorphism and folate/homocysteine concentrations in healthy young adults from Northern Ireland.
The SLC19A1 c.80G>A polymorphism was not strongly associated with either serum folate or homocysteine concentrations in either men or women. However, in women, but not in men, this polymorphism explained 5% of the variation in red blood cell (RBC) folate levels (P=0.02). Relative to women with the SLC19A1 c.80GG genotype, women with the GA and AA genotypes had higher RBC folate concentrations. Consequently, compared to women with the SLC19A1 c.80AA and GA genotypes, women who are homozygous for the 80G allele may be at increased risk of having a child affected with a neural tube defect and of developing pathologies that have been associated with folate insufficiency, such as cardiovascular disease.
Keywords: Reduced folate carrier, folate, homocysteine, RFC1, SLC19A1, SNP
Introduction
Folate/homocysteine metabolism supports several important biological processes including nucleic acid synthesis and the methylation of a variety of substrates (eg. proteins, DNA and lipids). A low folate, high homocysteine phenotype is a risk factor for, or a marker of, many human pathologies including spina bifida and cardiovascular disease (Lucock, 2000). Any impairment in folate bioavailability as a consequence of intake, transport or cellular utilization might contribute to the pathogenesis of diseases that have been associated with low folate status.
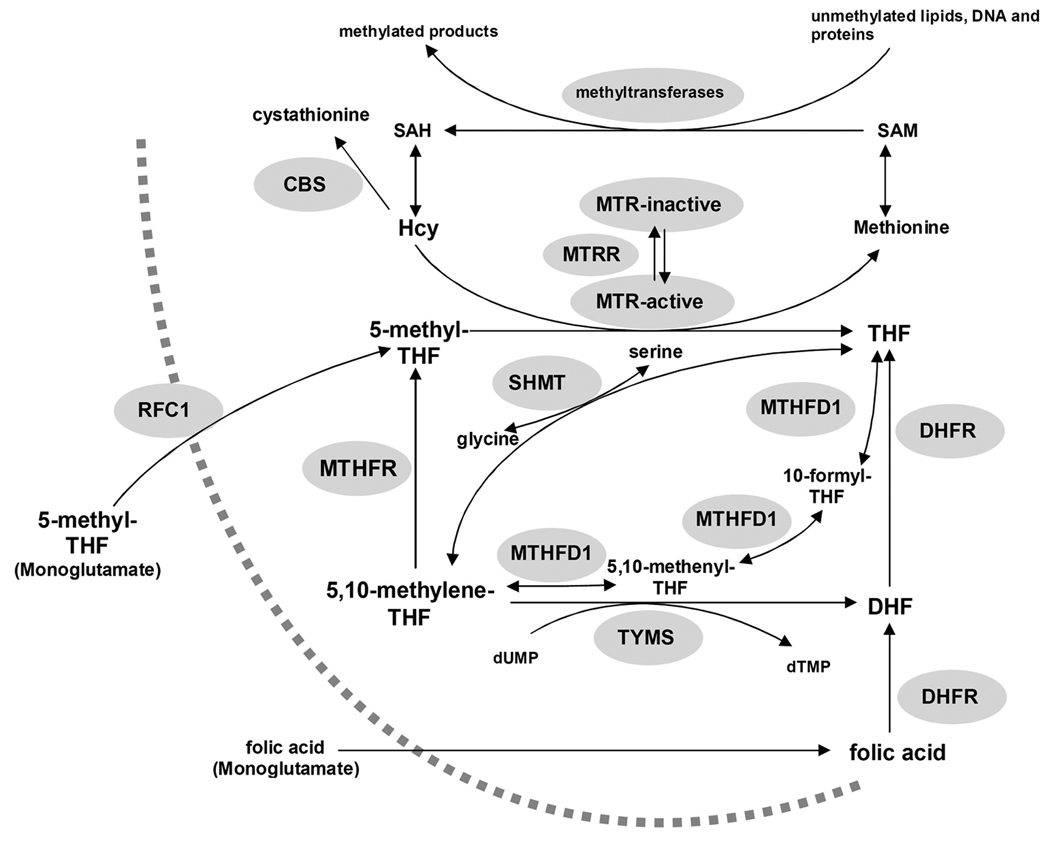
Folate transport is facilitated by the reduced folate carrier (MIM:600424; solute carrier family 19, gene names: SLC19A1, RFC1), organic anion carriers, folate receptors alpha and beta, and multidrug resistance-associated proteins (Matherly & Goldman, 2003). RFC1 is a high capacity, bi-directional transporter of 5-methyl-tetrahydrofolate (5-methylTHF) (Figure 1) and can also transport folic acid, albeit with lower capacity; it also facilitates transport of the antifolate drug methotrexate (MTX) (Matherly & Goldman, 2003). The SLC19A1 gene is polymorphic inhumans (Chango et al., 2000; Whetstine et al., 2002; http://snp500cancer.nci.nih.gov). The most extensively studied variant of the SLC19A1 gene is a single nucleotide polymorphism 80G>A in the coding region (SLC19A1 c.80G>A; rs61510559; often referred to as RFC1 80A>G), which results in the substitution of an arginine with a histidine at residue 27 in the amino acid sequence.
Figure 1.
Schematic representation of homocysteine/folate metabolism. CBS, cystathionine β-synthase; DHF, dihydrofolate; DHFR, dihydrofolate reductase; dTMP, deoxythymidine monophosphate; dUMP deoxyuridine monophosphate; Hcy homocysteine; MTHFD1, methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolaseformyltetrahydrofolate synthetase; MTHFR, 5,10-methylenetetrahydrofolate reductase; MTR, methionine synthase; MTRR, methionine synthase reductase; RFC1, Reduced folate carrier; SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine; SHMT, Serine hydroxymethyltransferase; THF, tetrahydrofolate; TYMS, thymidylate synthase.
The relationship between the SLC19A1 c.80G>A polymorphism and folate/homocysteine metabolism is unclear. Several groups have reported a lack of association with red blood cell (RBC) folate (Vesela et al., 2005; Chango et al., 2000), plasma folate (Winkelmayer et al., 2003; Chango et al., 2000; Devlin et al., 2006) and homocysteine (Winkelmayer et al., 2003; Yates & Lucock, 2005; Devlin et al., 2006). However, some studies have shown non-significant trends between the SLC19A1 c.80G>A polymorphism and RBC folate levels (Morin et al., 2003; Yates & Lucock, 2005), and among patients with thrombotic vascular disease increased plasma folate concentrations were observed in SLC19A1 c.80AA homozygotes (Yates & Lucock, 2005). In contrast, the largest study to date (N>10 000) identified a borderline significant decrease in serum folate levels in individuals with genotypes that included the SLC19A1 c.80A alleles; however, the relationship between the SLC19A1 genotype and RBC folate levels was not reported for that study (Fredriksen et al., 2007). Interestingly, two studies have provided evidence that the effect of the SLC19A1 c.80G>A variant may be influenced by the MTHFR g.677C>T genotype (MTHFR c.665C>T; rs1801133; often referred to as MTHFR 677C>T). Specifically, when SLC19A1 c.80G>A genotype was considered in the context of MTHFR g.677C>T genotype, doubly homozygous MTHFR g.677TT - SLC19A1 c.80GG individuals had elevated homocysteine (Chango et al., 2000; Devlin et al., 2006), relative to those with other MTHFR/SLC19A1 genotype combinations.
Taken together, the above reports suggest that further investigation of the relationship between the SLC19A1 c.80G>A polymorphism and folate/homocysteine phenotype is warranted. The aim of this study was to investigate the relationship between the SLC19A1 c.80G>A genotype and RBC folate, serum folate and homocysteine concentrations in young, reproductive age adults.
Materials and Methods
Study population
The Young Hearts Project (YH) is an ongoing longitudinal study designed to monitor cardiovascular disease risk factors in children and young adults living in Northern Ireland (Boreham et al., 1993). Briefly, a sample of 12 year old (n=509) and 15 year old (n=506) boys and girls were enrolled from post-primary schools in Northern Ireland between 1989 and 1990 (YH1). Between October 1997 and October 1999, all YH subjects were invited to participate in YH3, a hospital-based screening evaluation (Gallagher et al., 2002). The participation rate for YH3, which was conducted when the subjects were between 20 and 26 years of age, was 48.2% (n=489). Compared to non-respondents, YH3 subjects tended to be from families with higher socioeconomic status and to have had lower body mass indices at the baseline YH1 examination. In addition, male YH3 subjects were leaner and reported lower saturated fat intake at baseline relative to the male non-participants (Boreham et al., 2004). Ethical approval for each phase of the study was granted by the Research Ethics Committee of Queen’s University Belfast. The current paper is based on self-reported data on smoking status, use of alcohol and multivitamin supplements, and fasting blood samples collected as part of YH3.
Laboratory Methods
Blood samples were collected from fasted subjects for the determination of biochemical parameters and for DNA extraction (Miller et al., 1988). Homocysteine concentrations were measured by an established high performance liquid chromatography method (Ubbink et al., 1991). Serum folate concentrations were determined by time-resolved immunofluorescence on an AutoDelfia analyzer (Wallac, UK), and RBC folate concentrations were determined by a microbiological assay as previously described (Molloy & Scott, 1997), and are expressed as nanomoles per liter of packed RBCs.
SLC19A1 c.80A>G genotypes were determined by a modification of a published method (Skibola et al., 2004) using TaqMan 5’ Nuclease Real-Time PCR assay on a PTC-200 DNA Engine (Bio-Rad, Hercules, CA) with fluorescence detection by a Chromo4 Real-Time PCR Detector (Bio-Rad). Individual PCR amplification reactions (20 µl) were composed of 2 µl sample DNA, 1x TaqMan Universal PCR Master Mix, No AmpErase® UNG (Applied Biosystems, Foster City, CA), 0.5 µM forward primer (5’-GGCCTGACCCCGAGCT-3’) and 0.5 µM reverse primer (5’-AGCCGTAGAAGCAAAGGTAGCA-3’), 100 nM “G”-specific probe (VIC-CACGAGGCGCCGC), and 50 nM “A”-specific probe (6FAM-CGAGGTGCCGCCAG). The probes were synthesized by Applied Biosystems. PCR was performed with an initial incubation at 95°C for 10 min followed by 60 cycles of denaturation at 95°C for 30 sec and extension/5’ nuclease step at 63°C for 1 min. Dual fluorescence was detected after each completed 70 sec cycle. Genotypes were assigned using Opticon Monitor 3 analysis software (Bio-Rad). MTHFR g.677C>T genotypes have previously been reported (Kluijtmans et al., 2003).
Statistical methods
Descriptive analyses of the study variables were conducted using data from all study subjects, and included medians and percentiles for continuous variables (i.e. RBC folate, serum folate and homocysteine) and proportions for categorical variables (i.e. smoking status, use of alcohol and multivitamin supplements, and genotypes). Deviations from Hardy-Weinberg equilibrium for the SLC19A1 c.80G>A and MTHFR g.677C>T genotypes were assessed by χ2 analysis.
Simple and multiple linear regression analyses were conducted using log-transformed RBC folate, serum folate or homocysteine values as the main outcome measures. These analyses were restricted to the subset of YH3 study participants for whom there was complete data for all of the variables in any set of regression analyses. Simple linear regression models were fitted to the data and the coefficient of determination (R2) estimated from these models was used to assess the proportion of variation in the outcome variable that was explained by each predictor variable. SLC19A1 c.80G>A genotypes were coded using two dummy variables, one reflecting the comparison of the GG and GA genotypes and the other the comparison of the GG and AA genotypes. Each potential behavioral risk factor (i.e. use of multivitamins, cigarettes or alcohol) was coded as a dichotomous (i.e. yes/no) variable. Multiple regression models included predictor variables as well as two-way interaction terms. Interactions were coded as the product of a dichotomous behavioral variable and the dummy variables defining genotype. Hence, two interaction terms were included in each of the models that allowed for interactions. Nested models were compared to determine the change in the proportion of variation in the outcome variable explained by the addition of a single predictor variable or interaction term to the model. Specifically, the difference in the adjusted R2 values for a model with and a model without a given variable was calculated. The significance of individual predictor variables within a model was assessed using the t-statistic, and P-values<0.05 were considered to be statistically significant. All analyses were conducted using SAS version 9.1.
Results
The characteristics of YH3 study subjects are summarized, for the whole population and separately by sex, in Table 1. SLC19A1 c.80G>A and MTHFR g.677C>T genotypes in the population as a whole, and in the male and female subsets, were in Hardy-Weinberg equilibrium. Data from two study subjects, each of whom had an extreme outlying value for one of the biochemical measurements made in the YH3 samples (i.e. serum B12 = 1230 pmol/l and serum folate = 213 nmol/l), were excluded from all analyses.
Table 1.
Characteristics of Participants in the Young Hearts 3 Study.
| Variable | All (412)* | Males (N=225) | Females (N=186) |
|---|---|---|---|
| Biochemical Variables | |||
| Homocysteine (µmol/l) | |||
| N | 401 | 220 | 180 |
| Median (25th–75th percentile) | 8.9 (7.5–10.9) | 9.2 (7.7–11.0) | 8.6 (7.4–11.0) |
| RBC folate (nmol/l) | |||
| N | 364 | 193 | 170 |
| Median (25th–75th percentile) | 644.1 (479.0–844.8) | 704.8 (543.6–888.7) | 562.9 (428.7–764.7) |
| Serum folate (nmol/l) | |||
| N | 352 | 192 | 159 |
| Median (25th–75th percentile) | 12.8 (9.6–18.9) | 12.4 (9.4–18.9) | 13.2 (9.6–19.1) |
| Lifestyle Variables | |||
| Current use of cigarettes (N, %) | |||
| Yes | 156 (38.1) | 84 (37.7) | 72 (38.7) |
| No | 253 (61.9) | 139 (62.3) | 114 (61.3) |
| Current use of alcohol (N, %) | |||
| Yes | 336 (81.8) | 192 (85.3) | 144 (77.4) |
| No | 75 (18.3) | 33 (14.7) | 42 (22.6) |
| Current use of multivitamin supplements (N, %) |
|||
| Yes | 93 (22.6) | 44 (19.6) | 49 (26.3) |
| No | 318 (77.4) | 181 (80.4) | 137 (73.7) |
| Genotypes | |||
| SLC19A1 c.80G>A (N, %) | |||
| GG | 122 (29.6) | 67 (29.8) | 55 (29.6) |
| GA | 219 (53.2) | 123 (54.7) | 95 (51.1) |
| AA | 71 (17.2) | 35 (15.6) | 36 (19.4) |
| MTHFR g.677C>T (N, %) | |||
| CC | 178 (43.5) | 99 (44.4) | 78 (42.2) |
| CT | 176 (43.0) | 100 (44.8) | 76 (41.1) |
| TT | 55 (13.4) | 24 (10.8) | 31 (16.8) |
Information on sex was not available for one subject.
In the full study sample, SLC19A1 c.80G>A genotype was not a strong predictor of either serum folate (R2=0.004) or homocysteine concentrations and (R2=0.003), and was only modestly associated with RBC folate concentrations (R2=0.02, P=0.04) (Table 2). The median RBC folate concentrations of those with the SLC19A1 c.80AA, GA and GG genotypes were 699.8, 671.9 and 594.9 nmol/l RBCs, respectively.
Table 2.
Proportion of the variation (R2) in RBC folate, serum folate and homocysteine concentrations explained by SLC19A1 c.80G>A genotypes in YH3 study participants.
| All | Males | Females | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R2 (P-value)1 |
Variable Coefficient (SE)2 |
N | R2 (P-value)1 |
Variable Coefficient (SE)2 |
N | R2 (P-value)1 |
Variable Coefficient (SE)2 |
N | ||
| RBC Folate | ||||||||||
| SLC19A1 c.80G>A | 0.02 (0.04) | 363 | 0.01 (0.43) | 192 | 0.05 (0.02) | 169 | ||||
| GA | 0.05 (0.02) | 0.003 (0.03) | 0.09 (0.03) | |||||||
| AA | 0.07 (0.03) | 0.05 (0.04) | 0.10 (0.04) | |||||||
| Serum Folate | ||||||||||
| SLC19A1 c.80G>A | 0.004 (0.54) | 350 | 0.001 (0.95) | 191 | 0.01 (0.32) | 158 | ||||
| GA | 0.02 (0.03) | −0.01 (0.03) | 0.06 (0.04) | |||||||
| AA | 0.04 (0.04) | −0.10 (0.05) | 0.06 (0.05) | |||||||
| Homocysteine | ||||||||||
| SLC19A1 c.80G>A | 0.003 (0.53) | 400 | 0.01 (0.50) | 219 | 0.001 (0.89) | 179 | ||||
| GA | −0.02 (0.02) | −0.03 (0.02) | −0.01 (0.03) | |||||||
| AA | −0.02 (0.02) | −0.03 (0.03) | −0.003 (0.03) | |||||||
P-value for test of hypothesis of no linear association between the predictor variable and RBC folate, serum folate or homocysteine.
Estimated regression coefficients and standard errors. Variable coefficients estimate the change in RBC folate, serum folate or homocysteine concentration per unit change in the predictor variable.
As males and females differ with respect to several of the biochemical and lifestyle variables (Table 1) of interest in this study, further analyses were performed separately for males and females. Median values of the biochemical variables, by SLC19A1 c.80G>A genotype and sex, are provided in Table 3. The SLC19A1 c.80G>A genotype was not strongly associated with serum folate or homocysteine concentrations in either males or females (Table 2). Among females, RBC folate levels increased with the number of SLC19A1 c.80A alleles (Table 3), and the SLC19A1 c.80G>A genotype explained approximately 5% of the variation in RBC folate (P=0.02) (Table 2), Although RBC folate levels in males also increased with the number of SLC19A1 c.80A alleles (Table 3), SLC19A1 c.80G>A genotype explained a relatively small proportion of the variation (~1%) in RBC folates among males (Table 2).
Table 3.
Median RBC folate, serum folate and homocysteine values in subgroups of subjects defined by sex and RFC1 80G>A genotype.
| Biochemical Phenotype |
RFC1 80G>A genotype1 | |||||
|---|---|---|---|---|---|---|
| Males | Females | |||||
| GG | GA | AA | GG | GA | AA | |
| RBC folate (nmol/l) |
658.6 (55) | 715.5 (109) | 739.5 (29) | 481.1 (51) | 586.2 (85) | 624.1 (34) |
| Serum Folate (nmol/l) |
12.4 (59) | 12.4 (105) | 12.9 (28) | 12.1 (49) | 13.7 (81) | 13.5 (29) |
| Homocysteine (µmol/l) |
9.2 (65) | 8.9 (120) | 9.3 (35) | 9.0 (54) | 8.5 (90) | 9.5 (36) |
The number given in parentheses is the number of individuals.
Multivariate regression analyses were performed to estimate the proportion of RBC folate variation attributable to SLC19A1 c.80G>A genotype in the context of MTHFR g.677C>T genotype, an established determinant of folate/homocysteine phenotype (Jacques et al., 1996; Harmon et al., 1996; Kluijtmans et al., 2003). Analyses to assess potential interactions between the SLC19A1 c.80G>A and MTHFR g.677C>T genotypes were not undertaken due to the small number of subjects in several of the combined genotype categories. Accounting for the effects of the MTHFR g.677C>T variant, the SLC19A1 c.80G>A genotype accounted for approximately 3% of the variation in RBC folate levels in females, but less than 1% of the variation in RBC folate levels in males (Table 4). Further, among females, the SLC19A1 c.80G>A genotype accounted for a higher proportion of the variation in RBC folate than did the MTHFR g.677C>T variant (i.e. MTHFR g.677C>T genotype accounted for ~1% of the variation in RBC folate after accounting for the effects of the SLC19A1 c.80G>A genotype). No interactions between the SLC19A1 c.80G>A genotype and smoking, alcohol, or multivitamin use was observed in either males or females (data not presented), although it should be noted that firm conclusions are precluded by the small number of observations in several of the categories.
Table 4.
Summary of logistic regression modeling of log-RBC folate in Young Hearts 3 study participants.
| Model1 | Predictor Variables | Males | Females | |||||
|---|---|---|---|---|---|---|---|---|
| Variable Coefficient (SE) |
Adjusted R2 |
Change in Adjusted R2 |
Variable Coefficient (SE) |
Adjusted R2 | Change in Adjusted R2 |
|||
| RBC folate | ||||||||
| 1 | SLC19A1 c.80G>A | −0.0049 | 0.0734 | 0.0365 | 0.0143 | |||
| GA | 0.0002 (0.03) | 0.09 (0.03) | ||||||
| AA | 0.04 (0.04) | 0.10 (0.04) | ||||||
| 2 | MTHFR g.677C>T | 0.0706 | −0.0021 | 0.0175 | 0.0333 | |||
| CT | −0.06 (0.02) | −0.04 (0.03) | ||||||
| TT | −0.16 (0.04) | −0.09 (0.04) | ||||||
| 3 | MTHFR g.677C>T +SLC19A1 c.80G>A | 0.0685 | 0.0508 | |||||
| CT | −0.06 (0.02) | −0.04 (0.03) | ||||||
| TT | −0.16 (0.04) | −0.09 (0.04) | ||||||
| GA | 0.008 (0.03) | 0.09 (0.03) | ||||||
| AA | 0.05 (0.04) | 0.09 (0.04) | ||||||
Change in R2 was assessed relative to model 3.
Discussion
The data presented here indicate that in healthy young Northern Irish women the SLC19A1 c.80G>A polymorphism is significantly associated with RBC folate concentrations, with higher levels observed in women with genotypes including the A allele. While a similar trend was observed in men, this variant explained a smaller proportion of the variation in RBC folate in males as compared to females. The observed association between the SLC19A1 c.80A allele and relatively high RBC folate levels in women is consistent with the trend observed by Morin et al. (2003).
Neither in our study nor in those reported by others is there any apparent impact of the SLC19A1 c.80G>A polymorphism on homocysteine (Winkelmayer et al., 2003; Yates & Lucock, 2005; Devlin et al., 2006). Furthermore, our study provided no evidence that this polymorphism significantly influences variation in serum folate concentrations and in this respect it is similar to several other studies (Winkelmayer et al., 2003; Devlin et al., 2006; Chango et al., 2000), a notable exception being the very large (N= 10 601) study of Fredriksen et al. (2007) in which there was a borderline significant effect on serum folate.
Folic acid supplements prevent up to 70% of NTDs (MRC Vitamin Research Group, 1991; Czeizel & Dudas, 1992) and women carrying fetuses with spina bifida have low folate and high homocysteine concentrations (Mills et al., 1995; Kirke et al., 2004). The important role of RFC1 in folate transport has prompted several groups to evaluate the SLC19A1 c.80G>A polymorphism as a risk factor. Some groups have reported that the SLC19A1 c.80GG genotype is a risk factor for spina bifida (Shaw et al., 2002; De Marco et al., 2003; Pei et al., 2005; Morin et al., 2003), whilst others have found no evidence to support any such association (Relton et al., 2004; O'Leary V et al., 2006; Vieira et al., 2005). A single report has suggested that the SLC19A1 12 c.80AA genotype is a maternal risk factor for anencephaly, but not for spina bifida (Relton et al., 2003). Our observations are biologically consistent with the former reports.
Our findings may also have relevance to therapies involving the anti-folate drug methotrexate, which is transported to cells by RFC1. Others have reported that methotrexate concentrations (Laverdiere et al., 2002; Dervieux et al., 2004) are higher in patients with the SLC19A1 c.80AA genotype, suggesting that it might be possible to individually tailor dosing strategies by taking SLC19A1 c.80G>A genotype and sex into consideration.
In conclusion, we have demonstrated that the SLC19A1 c.80GG genotype is associated with relatively low folate concentrations in Northern Irish women. As a maternal low folate/high homocysteine phenotype is associated with increased risk of neural tube defects (NTDs) in offspring (Mills et al., 1995; Kirke et al., 2004), women with the SLC19A1 c.80GG genotype may have an increased risk of having a child affected by an NTD relative to those with the GA and AA genotypes. In addition, SLC19A1 c.80GG homozygous women may be at increased risk of a range of other major pathologies, including cardiovascular disease, in which a low folate/high homocysteine phenotype is a predisposing feature.
Acknowledgements
This work was supported by grants AR47663, ES013508, HD03915 and CA108862 from National Institutes of Health and by grant 4100038714 from the Pennsylvania Department of Health. Support for the Young Hearts Project was provided by the British Heart Foundation.
References
- Boreham C, Robson PJ, Gallagher AM, Cran GW, Savage JM, Murray LJ. Tracking of physical activity, fitness, body composition and diet from adolescence to young adulthood: The Young Hearts Project, Northern Ireland. Int J Behav Nutr Phys Act. 2004;1:14. doi: 10.1186/1479-5868-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boreham C, Savage JM, Primrose D, Cran G, Strain J. Coronary risk factors in schoolchildren. Arch Dis Child. 1993;68:182–186. doi: 10.1136/adc.68.2.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chango A, Emery-Fillon N, De Courcy GP, Lambert D, Pfister M, Rosenblatt DS, Nicolas JP. A polymorphism (80G->A) in the reduced folate carrier gene and its associations with folate status and homocysteinemia. Mol Genet Metab. 2000;70:310–315. doi: 10.1006/mgme.2000.3034. [DOI] [PubMed] [Google Scholar]
- Czeizel AE, Dudas I. Prevention of the first occurrence of neural-tube defects by periconceptional vitamin supplementation. N Engl J Med. 1992;327:1832–1835. doi: 10.1056/NEJM199212243272602. [DOI] [PubMed] [Google Scholar]
- De Marco P, Calevo MG, Moroni A, Merello E, Raso A, Finnell RH, Zhu H, Andreussi L, Cama A, Capra V. Reduced folate carrier polymorphism (80A-->G) and neural tube defects. Eur J Hum Genet. 2003;11:245–252. doi: 10.1038/sj.ejhg.5200946. [DOI] [PubMed] [Google Scholar]
- Dervieux T, Kremer J, Lein DO, Capps R, Barham R, Meyer G, Smith K, Caldwell J, Furst DE. Contribution of common polymorphisms in reduced folate carrier and gamma-glutamylhydrolase to methotrexate polyglutamate levels in patients with rheumatoid arthritis. Pharmacogenetics. 2004;14:733–739. doi: 10.1097/00008571-200411000-00004. [DOI] [PubMed] [Google Scholar]
- Devlin AM, Clarke R, Birks J, Evans JG, Halsted CH. Interactions among polymorphisms in folate-metabolizing genes and serum total homocysteine concentrations in a healthy elderly population. Am J Clin Nutr. 2006;83:708–713. doi: 10.1093/ajcn.83.3.708. [DOI] [PubMed] [Google Scholar]
- Fredriksen A, Meyer K, Ueland PM, Vollset SE, Grotmol T, Schneede J. Large-scale population-based metabolic phenotyping of thirteen genetic polymorphisms related to one-carbon metabolism. Hum Mutat. 2007;28:856–865. doi: 10.1002/humu.20522. [DOI] [PubMed] [Google Scholar]
- Gallagher AM, Savage JM, Murray LJ, Davey Smith G, Young IS, Robson PJ, Neville CE, Cran G, Strain JJ, Boreham CA. A longitudinal study through adolescence to adulthood: the Young Hearts Project, Northern Ireland. Public Health. 2002;116:332–340. doi: 10.1038/sj.ph.1900871. [DOI] [PubMed] [Google Scholar]
- Harmon DL, Woodside JV, Yarnell JW, Mcmaster D, Young IS, Mccrum EE, Gey KF, Whitehead AS, Evans AE. The common 'thermolabile' variant of methylene tetrahydrofolate reductase is a major determinant of mild hyperhomocysteinaemia. QJM. 1996;89:571–577. doi: 10.1093/qjmed/89.8.571. [DOI] [PubMed] [Google Scholar]
- http://snp500cancer.nci.nih.gov.
- Jacques PF, Bostom AG, Williams RR, Ellison RC, Eckfeldt JH, Rosenberg IH, Selhub J, Rozen R. Relation between folate status, a common mutation in methylenetetrahydrofolate reductase, and plasma homocysteine concentrations. Circulation. 1996;93:7–9. doi: 10.1161/01.cir.93.1.7. [DOI] [PubMed] [Google Scholar]
- Kirke PN, Mills JL, Molloy AM, Brody LC, O'leary VB, Daly L, Murray S, Conley M, Mayne PD, Smith O, Scott JM. Impact of the MTHFR C677T polymorphism on risk of neural tube defects: case-control study. BMJ. 2004;328:1535–1536. doi: 10.1136/bmj.38036.646030.EE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluijtmans LA, Young IS, Boreham CA, Murray L, Mcmaster D, Mcnulty H, Strain JJ, Mcpartlin J, Scott JM, Whitehead AS. Genetic and nutritional factors contributing to hyperhomocysteinemia in young adults. Blood. 2003;101:2483–2488. doi: 10.1182/blood.V101.7.2483. [DOI] [PubMed] [Google Scholar]
- Laverdiere C, Chiasson S, Costea I, Moghrabi A, Krajinovic M. Polymorphism G80A in the reduced folate carrier gene and its relationship to methotrexate plasma levels and outcome of childhood acute lymphoblastic leukemia. Blood. 2002;100:3832–3834. doi: 10.1182/blood.V100.10.3832. [DOI] [PubMed] [Google Scholar]
- Lucock M. Folic acid: nutritional biochemistry, molecular biology, and role in disease processes. Mol Genet Metab. 2000;71:121–138. doi: 10.1006/mgme.2000.3027. [DOI] [PubMed] [Google Scholar]
- Matherly LH, Goldman DI. Membrane transport of folates. Vitam Horm. 2003;66:403–456. doi: 10.1016/s0083-6729(03)01012-4. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JL, Mcpartlin JM, Kirke PN, Lee YJ, Conley MR, Weir DG, Scott JM. Homocysteine metabolism in pregnancies complicated by neural-tube defects. Lancet. 1995;345:149–151. doi: 10.1016/s0140-6736(95)90165-5. [DOI] [PubMed] [Google Scholar]
- Molloy AM, Scott JM. Microbiological assay for serum, plasma, and red cell folate using cryopreserved, microtiter plate method. Methods Enzymol. 1997;281:43–53. doi: 10.1016/s0076-6879(97)81007-5. [DOI] [PubMed] [Google Scholar]
- Morin I, Devlin AM, Leclerc D, Sabbaghian N, Halsted CH, Finnell R, Rozen R. Evaluation of genetic variants in the reduced folate carrier and in glutamate carboxypeptidase II for spina bifida risk. Mol Genet Metab. 2003;79:197–200. doi: 10.1016/s1096-7192(03)00086-6. [DOI] [PubMed] [Google Scholar]
- MRC Vitamin Study Research Group. Prevention of neural tube defects: results of the Medical Research Council Vitamin Study. Lancet. 1991;338:131–137. [PubMed] [Google Scholar]
- O'leary VB, Pangilinan F, Cox C, Parle-Mcdermott A, Conley M, Molloy AM, Kirke PN, Mills JL, Brody LC, Scott JM. Reduced folate carrier polymorphisms and neural tube defect risk. Mol Genet Metab. 2006;87:364–369. doi: 10.1016/j.ymgme.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Pei L, Zhu H, Ren A, Li Z, Hao L, Finnell RH. Reduced folate carrier gene is a risk factor for neural tube defects in a Chinese population. Birth Defects Res A Clin Mol Teratol. 2005;73:430–433. doi: 10.1002/bdra.20130. [DOI] [PubMed] [Google Scholar]
- Relton CL, Wilding CS, Jonas PA, Lynch SA, Tawn EJ, Burn J. Genetic susceptibility to neural tube defect pregnancy varies with offspring phenotype. Clin Genet. 2003;64:424–428. doi: 10.1034/j.1399-0004.2003.00161.x. [DOI] [PubMed] [Google Scholar]
- Relton CL, Wilding CS, Laffling AJ, Jonas PA, Burgess T, Binks K, Tawn EJ, Burn J. Low erythrocyte folate status and polymorphic variation in folate-related genes are associated with risk of neural tube defect pregnancy. Mol Genet Metab. 2004;81:273–281. doi: 10.1016/j.ymgme.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Shaw GM, Lammer EJ, Zhu H, Baker MW, Neri E, Finnell RH. Maternal periconceptional vitamin use, genetic variation of infant reduced folate carrier (A80G), and risk of spina bifida. Am J Med Genet. 2002;108:1–6. doi: 10.1002/ajmg.10195. [DOI] [PubMed] [Google Scholar]
- Skibola CF, Forrest MS, Coppede F, Agana L, Hubbard A, Smith MT, Bracci PM, Holly EA. Polymorphisms and haplotypes in folate-metabolizing genes and risk of non-Hodgkin lymphoma. Blood. 2004;104:2155–2162. doi: 10.1182/blood-2004-02-0557. [DOI] [PubMed] [Google Scholar]
- Ubbink JB, Hayward Vermaak WJ, Bissbort S. Rapid high-performance liquid chromatographic assay for total homocysteine levels in human serum. J Chromatogr. 1991;565:441–446. doi: 10.1016/0378-4347(91)80407-4. [DOI] [PubMed] [Google Scholar]
- Vesela K, Pavlikova M, Janosikova B, Andel M, Zvarova J, Hyanek J, Kozich V. Genetic determinants of folate status in Central Bohemia. Physiol Res. 2005;54:295–303. [PubMed] [Google Scholar]
- Vieira AR, Murray JC, Trembath D, Orioli IM, Castilla EE, Cooper ME, Marazita ML, Lennon-Graham F, Speer M. Studies of reduced folate carrier 1 (RFC1) A80G and 5,10-methylenetetrahydrofolate reductase (MTHFR) C677T polymorphisms with neural tube and orofacial cleft defects. Am J Med Genet A. 2005;135:220–223. doi: 10.1002/ajmg.a.30705. [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Witt TL, Matherly LH. The human reduced folate carrier gene is regulated by the AP2 and sp1 transcription factor families and a functional 61-base pair polymorphism. J Biol Chem. 2002;277:43873–43880. doi: 10.1074/jbc.M208296200. [DOI] [PubMed] [Google Scholar]
- Winkelmayer WC, Eberle C, Sunder-Plassmann G, Fodinger M. Effects of the glutamate carboxypeptidase II (GCP2 1561C>T) and reduced folate carrier (RFC1 80G>A) allelic variants on folate and total homocysteine levels in kidney transplant patients. Kidney Int. 2003;63:2280–2285. doi: 10.1046/j.1523-1755.2003.00025.x. [DOI] [PubMed] [Google Scholar]
- Yates Z, Lucock M. G80A reduced folate carrier SNP modulates cellular uptake of folate and affords protection against thrombosis via a non homocysteine related mechanism. Life Sci. 2005;77:2735–2742. doi: 10.1016/j.lfs.2005.02.029. [DOI] [PubMed] [Google Scholar]