Abstract
Bacterial species utilize a vast repertoire of surface structures to interact with their surroundings and employ a number of strategies to reconfigure the cellular envelope according to specific stimuli. Gram-positive bacteria, exemplified by Streptomyces and Bacillus species, control production of some exposed molecules by importing oligopeptide signals via permeases (Opp). Such oligopeptides modulate intracellular signaling pathways. In this work, we functionally characterized an Opp of the human pathogen Mycobacterium tuberculosis (Mtb) and propose its reannotation. Using genome-wide transcriptional profiling, we found that Opp was required to modulate (fold-change ranging from −3.5 to 2.0) the expression of several genes, most of them encoding surface-exposed molecules. These included the virulence-associated lipids mycolic acids and phthiocerol dimycocerosates (PDIMs) as well as PE-family proteins. By thin-layer chromatography and MALDI-TOF-MS we confirmed changes in the lipid profile, including an altered accumulation of triacylglycerides and an affected ratio of mycolic acids to PDIMs. An Opp loss of function mutant showed no in vitro growth defect, but had diminished burden during chronic infection and produced a slightly delayed time to death of animals when compared to WT Mtb infection.—Flores-Valdez, M. A., Morris, R. P., Laval, F., Daffé, M., Schoolnik, G. K. Mycobacterium tuberculosis modulates its cell surface via an oligopeptide permease (Opp) transport system.
Keywords: lipid virulence factors, cell wall modulation, chronic infection, surface interaction
Permease-mediated peptide transport mainly serves to facilitate the uptake of nutritionally favorable substrates. However, the ubiquitous presence of such cellular portals allows entry of specific peptide sequences that are able to transduce signals for adaptation to multiple environments. Sporulation, aerial hyphae formation, competence, and virulence are among the responses linked to specific peptide internalization. Such signaling often involves the participation of ion-linked and ATP-binding cassette transporters with oligopeptide-binding range. Small peptide-modulated signaling mechanisms have been proven only for Bacillus and Enterococcus species. In Bacillus subtilis, oligopeptide permease (Opp) is required to internalize the CSF and PhrE peptides, affecting the phosphatases RapA, RapB, RapC, and RapE and thus modulating the transcriptional activity of ComA and Spo0F (1,2,3). Bacillus thuringiensis Opp is required to modulate the global regulator PlcR (4). Likewise, in Bacillus cereus, the Opp-internalized small peptide PapR allows PlcR to bind to its DNA target (5). Enterococcus faecalis uses Opp to internalize cCF10, cAD1, and cPD1 to control transcription of genes and translation of transcripts required for mating (3, 6). Bioinformatic analysis to search for conserved phosphatase/signaling peptides modules, as are present in the Bacillus genome (2, 7), identified no candidate genes in Mycobacterium tuberculosis (Mtb).
Nevertheless, the Mtb genome contains 2 annotated permeases, for dipeptides (Dpp) and Opp, albeit doubts were cast about their true function (8). Therefore, we aimed to define their identity precisely and to provide a phenotypic characterization of responses to the deficiency of an Opp locus in Mtb, including the role of Opp during infection. The observed diminished fitness in murine macrophages of a Mycobacterium bovis BCG opp transposon-insertion mutant suggests a role in virulence (9). Furthermore, within an infecting pool constituted by 106 Mtb random transposon-insertion strains, opp mutants showed in vivo survival defects (10), a likely indicator of diminished virulence. Conversely, mycobacterial mutants in whiB3 (11) and sigma H (12), although they generate a bacterial burden indistinguishable from wild type (WT), elicit fatal infection in mice, thus constituting an exception to the rule that an in vivo survival defect is necessarily linked to virulence attenuation. Our results allow us to propose the reannotation of the Mtb Opp and Dpp systems, and show that Opp is required to modulate the ratio of 2 cell wall-associated lipids, phthiocerol dimycocerosates (PDIMs) and mycolic acids (Mycs), at both the transcript and lipid (end product) levels. In disagreement with previous in vivo observations where Opp was required for active infection (10), our results indicate that Opp was required for achieving WT-level burden during the chronic phase in an aerosol mouse model of infection, where the absence of a functional permease resulted in a slightly delayed time-to-death phenotype.
MATERIALS AND METHODS
Bacterial strains and media
The clinical Mtb isolate 1254 was used as the parental WT for construction of the permease knockout (KO) and complemented strains, and was cultured using Middlebrook 7H9 medium (BD, Franklin Lakes, NJ, USA), containing 10% Middlebrook oleic-acid-dextrose-catalase (OADC) enrichment (BD), 0.5% glycerol (Fisher Scientific, Pittsburgh, PA, USA) and 0.05% Tween 80 (Sigma-Aldrich, St. Louis, MO, USA), or 7H10 agar (BD) containing 10% OADC and 0.5% glycerol. Escherichia coli DH5α or TOP-10 (Invitrogen, Carlsbad, CA, USA) were used as hosts when constructing recombinant plasmids and were cultured using LB agar plates or broth. Hygromycin B (50 or 200 μg/ml, Invitrogen) and kanamycin (25 or 50 μg/ml, Sigma-Aldrich) were used when selecting for recombinant strains (Mtb) or plasmids (E. coli), respectively. When determining growth curves, the absorbance at optical density 600 nm (OD600) was recorded on a daily basis, and standard dilution and plating techniques were used to determine colony-forming units (CFUs), after 3 wk of incubation at 37°C.
Mutagenesis of the permease encoded by the Rv3665c-3663c genes in Mtb 1254
Specialized transduction (13) was employed to create a functional null mutant by targeted mutagenesis. The Rv3666c gene is predicted to be essential (14), and therefore the strategy employed served to leave this open reading frame and its associated promoter intact. The Rv3665c sequence until and including Rv3662c was deleted and marked with a cassette conferring hygromycin resistance. Briefly, a fragment upstream from the annotated dppB (Rv3665c) initiation codon, including the Rv3666c stop codon, was amplified using the primers dppB#7 (5′-GGTACCATTAATGGCGCTGTCGGCCGCCATCAAC-3′) and dppB#8 (5′-TCTAGAGGCGACTCGGCGCGCAACATACCAG-3′). In addition, the region downstream Rv3662c was amplified using the primers Rv3662#3 (5′-AAGCTTGGGCTGGCTGTTGCTGTGTTG-3′) and Rv3662#4 (5′-ACTAGTATTAATCACACAGTGGCCCGAGGAGGAA-3′). The sequences underscored are introduced restriction sites that facilitate directional cloning. A ΔRv3665c-Rv3662c::hyg allele was constructed by cloning these PCR products into pYUB854 (a gift of William R. Jacobs, Jr., Howard Hughes Medical Institute, Albert Einstein College of Medicine, Bronx, NY, USA) to create pRPM300. The temperature-sensitive mycobacteriophage phRP160 for transducing the ΔRv3665c-Rv3662c::hyg allele was generated by ligating PacI-digested (New England Biolabs, Ipswich, MA, USA) pRPM300 into the TM4-derived phAE87. All primers were synthesized and purchased at the Stanford Protein and Nucleic Acid Facility.
Mutagenesis of the permease encoded by the Rv1283c-Rv1280c genes in Mtb 1254
We followed a similar approach to delete the other annotated permease system. The Rv1283c-Rv1280c genes were deleted and marked with a cassette conferring hygromycin resistance. Briefly, a fragment upstream from the permease locus was amplified using the primers oppB#1 (5′-GGTACCATTAATGCTTGAAGTCGTCGTCGGTGAAA-3′) and oppB#2 (5′-TCTAGAGCCAGCAGCACCAGGTAGTTGAG-3′). In addition, a region downstream from the permease locus was amplified using the primers oppA#3 (5′-AAGCTTCTGGCAGACCTGGACTACACC-3′) and oppA#4 (5′-ACTAGTATTAATTTGTTTGCCGCCGAGAAGC-3′). The sequences underscored are restriction sites introduced to facilitate directional cloning. A ΔRv3665c-Rv3662c::hyg allele was constructed to create pRPM310 and used to generate the temperature-sensitive, KO delivery vector, mycobacteriophage phRP170.
Confirmation of mutated permease alleles in Mtb 1254
Hygromycin-resistant transductants were picked and cultivated in liquid 7H9 broth with hygromycin. Crude DNA-containing extracts were prepared from well-grown heat-killed cultures. Briefly, a 1-ml sample of culture in a 2-ml screw-cap tube was placed in a boiling water bath for 30 min and then centrifuged at maximum speed for 10 min, and a sample of supernatant was removed for analysis by PCR. Transductants were confirmed by the presence of a specific PCR amplicon with a primer targeting the hygromycin-resistant cassette (5′-AACTGCTCGCCTTCACCTTCCT-3′) and primers targeting downstream from the respective permease locus (dpp#6 5′-AAGCTTGAGTTTAGGAACTACTCATCC-3′ and opp#6 (5′-AAGCTTTGAGCTGGTCGCCGCCGGCTC-3′).
Functional characterization of the Opp system
Mtb 1254 strains were cultured in 7H9 broth to an OD600 of 1, and a 5-μl drop of each strain was inoculated onto plates with or without 5 μg/ml of the antibiotic phosphinothricin tripeptide (bialaphos, a gift of Prof. Charles Thompson, University of British Columbia, Vancouver, BC, Canada). The plates were incubated at 37°C for 3–4 wk before reading them and recording the effect of exposure to bialaphos. Nonfunctional mutations in the Opp confer resistance to bialaphos because of a failure to internalize the bioactive principle phosphinothricin.
Complementation of the opp KO
A 7.6-kb fragment, containing 482 bp upstream of the Rv3666c start codon and 1353 bp downstream of the Rv3662c stop codon (coordinates 4099912–4107566 of the H37Rv genome; TubercuList: http://genolist.pasteur.fr/TubercuList/) was obtained by Pci I (New England Biolabs) digestion of pI568 (kindly provided by Dr. Roland Brosch, Pasteur Institute, Paris, France) and subsequently cloned into the same site of the integrative vector pMV361 (8) to create pMF-Rv3666c-Rv3662c. The identity of the new plasmid was confirmed both by restriction digestion and sequencing. Transformants were selected after 4 wk of growth at 37°C on plates with hygromycin and kanamycin, to produce M. tuberculosis ΔRv3665c- Rv3662c::hyg attB:: pMF-Rv3666c-Rv3662c (complemented).
Microarray experiments
Mtb strain 1254 WT and its isogenic Δopp KO and complemented strains were cultured in liquid medium, in a 150-ml volume, contained in roller bottles, and incubated at 37°C. When cells reached OD600 of 0.3, 1.0, and 2.0, an aliquot of 30, 10, and 10 ml, respectively, was taken and centrifuged 3 min at 3000 g at room temperature, and the supernatant was discarded, with cell pellets immediately frozen by inclusion into dry ice. Frozen cell pellets were stored at −80°C for later RNA isolation. In all experiments, 2–3 μg of total RNA from WT was labeled with Cy5, and RNA from the opp KO was labeled with Cy3, using SuperScript III (Invitrogen) or AffinityScript (Stratagene, La Jolla, CA, USA) following the manufacturer’s recommendations. Here 70-mer oligonucleotide microarrays (Operon Technologies, Huntsville, AL, USA) were first prehybridized for 1 h in 5× SSC, 1% BSA, and 0.1% SDS and washed with H2O and isopropanol. After the prehybridization, 10 μl of hybridization solution (labeled cDNA, 5 μg tRNA, 2× SSC, 25% formamide, and 0.1% SDS; all chemicals from Sigma-Aldrich) were hybridized at 54°C for 16–20 h. Microarrays were scanned by using GenePix 4000A (Axon Instruments-Molecular Devices, Sunnyvale, CA, USA). Fluorescence intensities of the 2 channels at each spot were quantified by using the GenePix software. After data for each array were normalized using a computed normalization procedure (Stanford Microarray Database, Stanford University), expression ratios were calculated from 2 or 3 biological duplicates and 2 microarrays for each biological replicate. Furthermore, the experiment was repeated on a different date, and all data were scored for statistical significance by using significance analysis of microarrays (SAM; 9) with a false discovery rate (FDR) of 0%, performed on genes with no flag in the spotted arrays and whose regression correlation was ≥0.6. Genes whose differential expression was ≥1.5-fold between strains at any particular growth phase analyzed were further considered for organization according to functional categories, as described in the annotated Mtb genomes or based on in silico prediction of their function. Microarray images were created using TIGR MeV 4.1 v.10.2 (http://www.tm4.org/mev.html). All whole-transcriptome data were deposited at Stanford Microarray Database (http://smd. stanford.edu/cgi-bin/tools/display/listMicroArrayData.pl?table Name=publication), and data will also be publicly available at Gene Expression Omnibus (GEO; accession no. GSE18426) and ArrayExpress.
In silico function prediction for genes differentially expressed between strains
Protein sequences translated in silico from genes whose expression was significantly different between samples were obtained from the Mtb H37Rv genome sequence available at TubercuList and used to predict their function by comparison with nonredundant protein sequence databases from the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi) using Position-Specific Iterated BLAST (PSI-BLAST) and the normal mode of the Simple Modular Architecture Research Tool (SMART, http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1). Protein family characteristics were checked at Pfam (http://pfam.sanger.ac.uk).
Extraction and purification of mycobacterial lipids
Lipids were extracted from cell pellets first with CHCl3:CH3OH (Scharlau-Chemie, Barcelona, Spain) (1:2 v/v) for 24 h at room temperature, and twice with CHCl3:CH3OH (2:1 v/v) for 2 d. Crude extracts were washed twice with distilled water and evaporated to dryness.
Analytical procedures for lipid analysis
The lipid extracts were analyzed by thin-layer chromatography on silica gel Durasil 25-precoated plates (0.25 mm thickness; Macherey-Nagel, Hoerd, France) using petroleum ether/diethyl ether (Scharlau-Chemie) (90:10 v/v) as eluent. Revelation of lipids compounds was performed by spraying the plates with phosphomolybdic acid hydrate (Sigma-Aldrich) (10% in ethanol, Scharlau-Chemie), followed by charring.
Alkaline hydrolysis of lipids was performed with 1 M sodium methanolate for 1 h at 37°C. After neutralization with glacial acetic acid, the mixture was dried under a stream of nitrogen, and then lipids were extracted with diethyl ether and washed twice with water. The resulting products were analyzed by matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry.
Spectrometric method
MALDI-MS detection of samples in reflectron mode was performed on a 4700 Proteomics Analyzer (Voyager DE-STR; Applied Biosystems, Framingham, MA, USA) equipped with a Nd:YAG laser (355 nm) operating by pulses of 500 ps with a frequency of 200 Hz. We accumulated 2500 shots in positive ion mode, and mass spectrometry data were acquired using the instrument default calibration. Lipid samples were dissolved in chloroform and were directly spotted onto the target plate as 0.5-μl droplets, followed by the addition of 0.5 μl of matrix solution. Samples were allowed to crystallize at room temperature. The matrix used was 2,5-dihydroxybenzoic acid (Sigma-Aldrich, Lyon, France) (10 mg/ml) in CHCl3/CH3OH (1:1 v/v).
Growth and virulence assessment in mice
Animal protocols were approved by the Johns Hopkins University Animal Care and Use Committee. For single-mutant infections, 3 groups of 6-wk-old BALB/c mice (Charles River, Frederick, MD, USA) were infected using a Middlebrook inhalation exposure system (Glas-Col, Terre Haute, IN, USA) with 10 μl of a midlog phase Mtb culture diluted to OD600 ∼ 0.08. For mixed infection, 24 BALB/c mice were infected with an aerosolized 1:1 mixture of WT1254:opp KO under similar conditions.
Four mice from each group were weighed and sacrificed at d 1, 7, 14, 28, 84, and 112 postinfection to determine the number of bacilli present in the lung and spleen. Mouse organs were aseptically removed, homogenized by bead beating, and serially diluted. Appropriate dilutions were plated onto Middlebrook 7H10 selective agar plates to determine the number of CFUs present. Statistical significance (P≤0.05) for both in vivo and in vitro CFU counts was analyzed using Microsoft Excel 2004 for Mac (Microsoft Corporation, Redmond, WA, USA) 2-tailed Student’s t test, after conducting an ANOVA test to determine the quality (equal/unequal) of data variance. Eleven mice from each group were allocated to assess the time required to reach mortality (time to death). For histological analysis, representative tissue samples from each group were fixed in 10% formaldehyde, paraffinized, sectioned, and stained with hematoxylin-eosin using standard procedures. GraphPad Prism (GraphPad Software, La Jolla, CA, USA) was used to construct and compare survival curves, whose statistical significance (P≤0.05) was assessed by the Logrank test to compare Kaplan-Meier curves.
RESULTS
Mtb Rv3666c-Rv3663c encodes an Opp transport system dispensable for in vitro growth
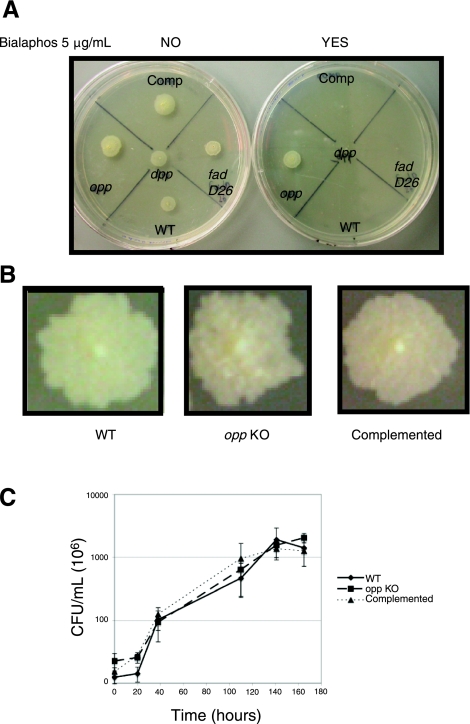
The annotated genomic sequences of Mtb H37Rv and CDC1551 contain 2 predicted peptide permease operons homologous to ubiquitous dpp (Rv3666c-Rv3663c; MT3767-MT3764) and opp (Rv1283c-Rv1280c; MT1320-MT1317) (15, 16). Recently, in M. bovis var. BCG, experimental evidence suggested the Rv1283c-Rv1280c operon was an Opp, albeit arguments suggesting a possible misannotation were presented (8). Therefore, we aimed to precisely define which cluster of Mtb genes corresponds to each function. For this, a biochemical susceptibility test using the toxic tripeptide bialaphos (17) was employed (1, 2, 18,19,20,21). WT Mtb proved susceptible to bialaphos, whereas the ΔRv3665c-Rv3663c-KO mutant exhibited normal growth in the presence or absence of the toxic tripeptide. Reintroducing a functional copy of the permease into the null mutant restored sensitivity to bialaphos (Fig. 1A). The ΔRv1283c-Rv1280c-KO mutant was susceptible as well as the control fadD26 (PDIM−) strain (Fig. 1A). Based on these results and the arguments provided by Greene et al. (8), we concluded that the Rv3666c-Rv3663c locus encodes an Opp transport system, whereas the Rv1283c-Rv1280c locus encodes a Dpp system. Deletion of the Opp system produced rugose colonies that were more wrinkled than WT. This morphological phenotype reverted to WT-like on complementation (Fig. 1B). However, the lack of the Opp system results in no significant difference during in vitro growth in liquid medium compared to WT and complemented strains (Fig. 1C).
Figure 1.
Functional characterization and reannotation of the M. tuberculosis oligopeptide (Opp) transport system. A) WT M. tuberculosis and the dpp, PDIM− (fadD26), and opp mutants, including the complemented (Comp) opp mutant, were cultured on solid medium in the absence (left plate, NO) or presence (right plate, YES) of bialaphos at a 5 μg/ml concentration, and incubated at 37°C for 3 wk. B) WT, opp mutant (opp KO), and its complemented strains were plated for single colonies on solid medium and incubated for 7 wk at 37°C. C) Growth curve (CFU/ml) of WT, opp KO, and complemented strains.
Disruption of the opp locus affects the expression of genes involved in the production of mycobacterial lipids
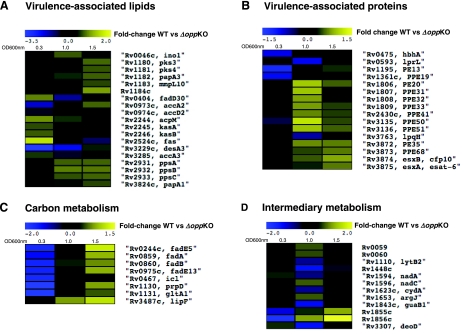
A genome-wide portrait of effects occurring within the transcriptome belonging to the opp KO mutant was obtained by comparing the expression profiles to that of the WT strain. Three different growth phases were examined: midlog (OD600∼0.3), log to stationary phase transition (OD600∼1), and stationary phase (OD600∼2.0; Fig. 1C–D). Our experiments showed that Opp-modulated genes clustered in different categories: 1) genes known (acpM, kasA, kasB, fas, accA3, and desA3) or predicted (fadD30, accA2, accD2) to be required for biosynthesis of mycolic acids (Fig. 2A; refs. 22, 23); 2) genes required for biosynthesis of the complex lipids PDIMs (ppsA, ppsB, and ppsC; refs. 24,25,26,27,28) and polyacyltrehaloses (pks3-pks4=msl3; refs. 27,28,29) (Fig. 2A); 3) genes required to produce virulence-associated proteins, such as the 19-kDa antigen (lpqH) (30, 31), cfp10-esat6 (32, 33) (Fig. 2B), the heparin-binding hemagglutinin (hbhA), PE13, and PPE19 (34, 35); 4) genes involved in carbon metabolism (β-oxidation of fatty acids fadE5, fadE13, fadA) (36, 37), and methyl citrate/glyoxylate cycle (icl, prpD, gltA1) (36, 37), as well as lipF (15) (Fig. 2C); and 5) genes required for intermediary metabolism (38,39,40) (lytB2, guaB1, deoD; Fig. 2D). The described differences varied in magnitude with respect to the growth phase analyzed. In some cases, increasing (MT2466, MT2467, MT3412, MT3413, fadE5, icl, fadA, accA2, fadE13, prpD, gltA1, Rv1184c, PE13, Rv1855c, Rv1856c, modA) or decreasing expression in the WT compared to the opp KO mutant. Other genes displayed a log-to-stationary phase up- or down-regulation, mostly within the “intermediary metabolism” category (Rv0059, Rv0060, lytB2, Rv1448c, nadA, cydA, guaB1, and deoD), with ino1, lprL, and lpqH not belonging to this group. Detailed expression values of significantly affected genes at each growth phase are shown in Table 1.
Figure 2.
Regulation of genes encoding proteins predicted to be involved in PDIMs, mycolic acids, PPE-family, ESAT-6/CFP-10, carbon, and intermediary metabolism. Yellow-blue display summarizes the regulation of selected Opp-modulated genes, comparing WT vs. mutant (opp KO) expression. cDNA ratios of statistically significantly affected genes were averaged, log2 transformed, and displayed according to the color code at the top of the display. Black fields indicate genes with no significant change at a particular growth phase. Experimental conditions were as indicated in Materials and Methods. Genes were selected based on a demonstrated or presumed function of the encoded proteins and grouped in the following categories: virulence-associated lipids (A) and proteins (B) or involved in carbon (C) or intermediary metabolism (D).
TABLE 1.
Gene expression ratios for genes differentially expressed in M. tuberculosis WT compared to opp KO mutant
| Rv or MT numbera | Fold change WT/opp KO OD600 0.3 | Fold change WT/opp KO OD600 1.0 | Fold change WT/opp KO OD600 2.0 | Annotationb or similarityc to protein domains |
|---|---|---|---|---|
| MT0719.1 | 1.3 | 0.6 | 1.3 | HP, 62 aa |
| MT1178 | 0.6 | 0.9 | 1.4 | HP, 51 aa |
| MT1585.1 | 1.0 | 1.8 | 1.4 | HP 55 aa |
| MT1676 | 1.2 | 1.3 | 1.6 | CHP similar to PspA/IM30 |
| MT2466 | 1.0 | 1.6 | 2.0 | HP, 71 aa, PhoP-dependent |
| MT2467 | 1.1 | 2.1 | 2.3 | HP, 54 aa |
| MT3220.2 | 1.0 | 1.9 | 1.5 | HP, 81 aa |
| MT3412 | 1.0 | 1.4 | 1.9 | HP, 56 aa |
| MT3413 | 1.1 | 1.5 | 2.3 | Rv3312A |
| MT3972.1 | 1.1 | 2.2 | 0.9 | HP, 46 aa |
| Rv0046c | 0.8 | 1.7 | 0.9 | Ino1 |
| Rv0059 | 1.1 | 1.6 | 1.0 | HP, 230 aa |
| Rv0060 | 1.0 | 1.6 | 1.0 | CHP, high-affinity ADP-ribose binding module |
| Rv0082 | 1.1 | 1.7 | 1.3 | Probable oxidoreductase |
| Rv0242c | 0.9 | 1.0 | 0.6 | FabG4 |
| Rv0244c | 0.3 | 1.1 | 2.4 | FadE5 |
| Rv0290 | 1.0 | 0.6 | 1.1 | Probable conserved transmembrane protein |
| Rv0341 | 1.1 | 0.8 | 0.6 | IniB |
| Rv0404 | 1.9 | 0.4 | 0.9 | FadD30 |
| Rv0440 | 0.7 | 1.6 | 1.0 | GroEL2 |
| Rv0467 | 0.3 | 0.9 | 1.1 | Icl |
| Rv0475 | 0.6 | 1.1 | 1.0 | HbhA |
| Rv0508 | 1.0 | 0.6 | 0.9 | CHP similar to thiol-disulfide isomerase |
| Rv0509 | 1.3 | 0.5 | 1.1 | HemA |
| Rv0583c | 1.6 | 0.5 | 0.9 | LpqN |
| Rv0593c | 1.1 | 0.5 | 1.0 | LprL |
| Rv0595c | 1.3 | 0.6 | 1.1 | CHP, PIN domain |
| Rv0651 | 1.1 | 1.6 | 0.8 | RplJ |
| Rv0693 | 1.4 | 0.5 | 1.2 | PqqE |
| Rv0700 | 1.1 | 1.6 | 1.1 | NusE |
| Rv0702 | 1.1 | 1.6 | 1.0 | RplD |
| Rv0704 | 1.2 | 1.6 | 1.1 | RplB |
| Rv0706 | 1.2 | 1.9 | 1.3 | RplV |
| Rv0708 | 1.1 | 1.6 | 1.1 | RplP |
| Rv0710 | 1.1 | 1.6 | 1.1 | RpsQ |
| Rv0714 | 1.1 | 1.6 | 1.2 | RplN |
| Rv0716 | 1.1 | 1.6 | 1.1 | RplE |
| Rv0722 | 1.1 | 1.6 | 1.1 | RpmD |
| Rv0859 | 0.4 | 1.2 | 1.8 | FadA |
| Rv0860 | 0.3 | 1.3 | 1.6 | FadB |
| Rv0878c | 2.2 | 1.9 | 1.7 | PPE13 |
| Rv0963c | 0.7 | 0.6 | 0.7 | CHP similar to Streptomyces secreted protein |
| Rv0973c | 0.3 | 1.3 | 1.7 | AccA2 |
| Rv0974c | 0.6 | 1.0 | 1.1 | AccD2 |
| Rv0975c | 0.4 | 1.1 | 1.8 | FadE13 |
| Rv1110 | 1.3 | 0.6 | 1.1 | LytB2 |
| Rv1130 | 0.3 | 0.9 | 1.6 | PrpD |
| Rv1131 | 0.3 | 0.9 | 1.4 | GltA1 |
| Rv1180 | 1.0 | 1.0 | 1.7 | Pks3 |
| Rv1181 | 0.9 | 0.9 | 1.6 | Pks4 |
| Rv1182 | 1.0 | 1.4 | 1.7 | PapA3 |
| Rv1183 | 0.9 | 0.9 | 1.6 | MmpL10 |
| Rv1184c | 1.0 | 1.3 | 2.1 | Similar to PE proteins |
| Rv1195 | 0.4 | 0.9 | 1.4 | PE13 |
| Rv1258c | 0.9 | 0.9 | 0.6 | Probable conserved integral membrane transport protein |
| Rv1361c | 0.5 | 0.9 | 1.0 | PPE19 |
| Rv1448c | 1.1 | 0.4 | 0.8 | Probable transaldolase |
| Rv1535 | 1.1 | 1.9 | 1.2 | HP, 78 aa |
| Rv1594 | 1.4 | 0.6 | 1.0 | NadA |
| Rv1596 | 1.3 | 1.7 | 1.0 | NadC |
| Rv1623c | 0.9 | 0.6 | 0.9 | CydA |
| Rv1642 | 1.0 | 1.6 | 1.4 | RpmI |
| Rv1653 | 1.3 | 1.6 | 0.9 | ArgJ |
| Rv1687c | 1.0 | 0.6 | 1.0 | Probable conserved ATP-binding transporter |
| Rv1806 | 1.0 | 2.0 | 1.5 | PE20 |
| Rv1807 | 1.2 | 1.9 | 1.2 | PPE31 |
| Rv1808 | 1.0 | 1.7 | 1.2 | PPE32 |
| Rv1809 | 1.0 | 1.7 | 1.4 | PPE33 |
| Rv1810 | 0.9 | 1.4 | 1.7 | CHP similar to LprJ |
| Rv1843c | 1.1 | 0.5 | 1.0 | GuaB1 |
| Rv1855c | 0.5 | 1.4 | 2.2 | Possible tetrahydromethanopterin reductase |
| Rv1856c | 0.3 | 2.1 | 4.3 | Possible short-chain dehydrogenase |
| Rv1857 | 0.4 | 1.5 | 2.2 | ModA |
| Rv2023c | 1.0 | 0.6 | 1.0 | HP, 119 aa |
| Rv2055c | 0.9 | 1.4 | 1.6 | RpsR2 |
| Rv2056c | 0.9 | 1.4 | 1.7 | RpsN2 |
| Rv2057c | 0.9 | 1.5 | 1.8 | RpmG1 |
| Rv2147c | 1.2 | 1.7 | 1.1 | CHP similar to L. plantarum putative cell division protein |
| Rv2160c | 1.1 | 0.5 | 1.1 | CHP, similar to G. violaceous TetR-family transcriptional regulatory protein |
| Rv2243 | 1.9 | 1.5 | 1.2 | FabD |
| Rv2244 | 2.0 | 1.5 | 1.2 | AcpM |
| Rv2245 | 1.6 | 1.0 | 1.0 | KasA |
| Rv2246 | 1.7 | 1.2 | 1.3 | KasB |
| Rv2253 | 1.2 | 1.7 | 1.1 | Possible secreted unknown protein |
| Rv2280 | 1.1 | 0.6 | 0.9 | Probable dehydrogenase |
| Rv2369c | 1.4 | 1.3 | 1.6 | HP, 100 aa |
| Rv2430c | 1.0 | 1.6 | 1.4 | PPE41 |
| Rv2524c | 2.7 | 1.2 | 0.5 | Fas |
| Rv2542 | 1.2 | 0.6 | 1.1 | CHP, α/β hydrolase fold |
| Rv2557 | 1.2 | 0.4 | 1.4 | CHP, thought to be involved in the persistence in the host |
| Rv2616 | 1.1 | 0.6 | 1.0 | CHP similar to C. glutamicum GTPase |
| Rv2724c | 0.5 | 1.3 | 1.4 | FadE20 |
| Rv2784c | 1.1 | 1.6 | 1.2 | LppU |
| Rv2821c | 1.1 | 0.6 | 0.9 | CHP, CRISPR-associated protein |
| Rv2840c | 1.1 | 1.6 | 1.5 | CHP, COG2740 implicated in transcription antitermination |
| Rv2846c | 1.6 | 1.5 | 1.9 | EfpA |
| Rv2931 | 1.2 | 1.7 | 1.7 | PpsA |
| Rv2932 | 1.3 | 1.8 | 1.8 | PpsB |
| Rv2933 | 1.1 | 1.6 | 1.7 | PpsC |
| Rv2987c | 1.0 | 1.6 | 0.5 | LeuD |
| Rv2989 | 1.1 | 1.6 | 0.6 | Probable IclR-family transcriptional regulator. |
| Rv2990c | 0.9 | 0.6 | 1.3 | HP, SAM-dependent methyltransferase family protein |
| Rv3135 | 0.8 | 2.2 | 1.4 | PPE50 |
| Rv3136 | 1.0 | 1.9 | 1.5 | PPE51 |
| Rv3229c | 0.1 | 0.2 | 1.4 | DesA3 |
| Rv3230c | 0.2 | 0.3 | 0.8 | Hypothetical oxidoreductase |
| Rv3285 | 1.5 | 0.6 | 1.1 | AccA3 |
| Rv3307 | 1.5 | 0.5 | 1.0 | DeoD |
| Rv3414c | 1.5 | 1.7 | 1.3 | SigD |
| Rv3418c | 0.9 | 1.8 | 1.4 | GroES |
| Rv3460c | 1.1 | 1.7 | 1.3 | RpsM |
| Rv3462c | 1.2 | 1.6 | 1.3 | InfA |
| Rv3487c | 1.0 | 1.7 | 2.2 | LipF |
| Rv3763 | 1.2 | 0.6 | 1.1 | LpqH |
| Rv3788 | 1.2 | 1.6 | 1.1 | HP, GreA/GreB domain |
| Rv3822 | 1.1 | 1.4 | 1.8 | CHP, COG3008, paraquat-inducible protein |
| Rv3824c | 1.0 | 1.4 | 1.6 | PapA1 |
| Rv3862c | 1.3 | 1.5 | 1.5 | WhiB6 |
| Rv3872 | 1.1 | 1.7 | 1.6 | PE35 |
| Rv3873 | 1.1 | 1.4 | 1.6 | PPE68 |
| Rv3874 | 1.2 | 1.5 | 1.8 | EsxB, CFP-10 |
| Rv3875 | 1.1 | 1.6 | 1.6 | EsxA, ESAT-6 |
Gene induction ratios were calculated from the comparison with RNA of each strain isolated at the same growth phase, as stated in the Materials and Methods section.
According to Cole et al. (15) and database (http://genolist.pasteur.fr/TubercuList/ or http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?database=gmt).
nnotation corresponding to Cole et al. (15) and database (http://genolist.pasteur.fr/TubercuList/).
Similarity as predicted by tools (http://blast.ncbi.nlm.nih.gov/Blast.cgi and http://smart.embl-heidelberg.de/smart/set_mode. cgi?NORMAL=1).
Growth-phase-dependent content of PDIMs, Mycs, and triacylglycerides (TAGs) is deregulated in the opp KO mutant
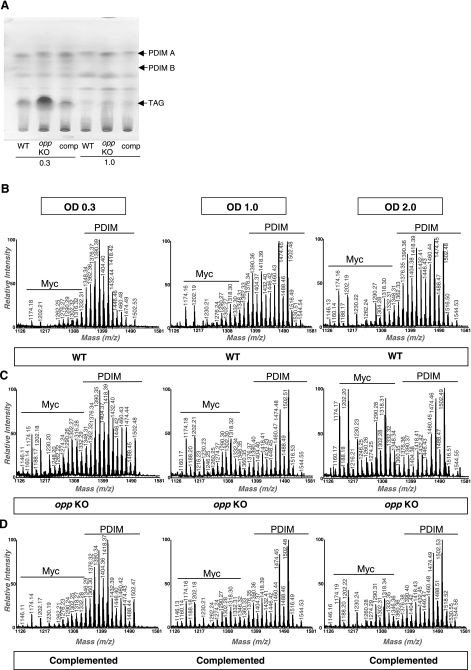
To confirm transcriptional results further, the levels of 2 types of well-known and relevant Mtb protein antigens were assessed: Western blot showed that secretion of ESAT-6 and CFP-10 was not affected by the presence or absence of Opp (data not shown), whereas thin-layer chromatography and MALDI-TOF mass spectroscopy indicated that the absence of Opp resulted in augmented accumulation of TAGs (Fig. 3A) and in deregulation of the growth-stage-dependent PDIM/Myc ratio (Fig. 3B–D). Changes were not detected for any other lipid species, including polyacyltrehaloses and diacyltrehaloses. The series of pseudomolecular ion (M+Na+) peaks that were observed at 1174, 1202, 1290, and 1318 m/z corresponded to mycolates (41), whereas the prominent peaks at 1348 through 1502 m/z corresponded to PDIMs with 89–100 carbon atoms (42). Based on peak intensity estimations, and as the culture progressed, we observed that the PDIM/Myc ratio was variable in the WT in a growth-phase-dependent manner, with a gradual increase of the Myc content relative to PDIMs set as reference (Fig. 3B). This capacity of slightly increasing Myc content was lost in the opp KO mutant, which did not decrease its PDIM/Myc ratio until late in stationary phase (OD 2.0) and which possessed higher Myc relative intensities compared to WT cells (Fig. 3C). Complementation of the mutant resulted in a WT-like PDIM/Myc ratio with recovered capacity to vary according to growth phase (Fig. 3D).
Figure 3.
PDIM biosynthesis, Myc biosynthesis, and TAGs are modulated via Opp. A) TLC analyses of lipids extracted from the M. tuberculosis WT, opp mutant (opp KO), and complemented (comp) strains. Arrows indicate the positions of PDIM-A, PDIM-B, and triacylglycerol (TAG). B–D) MALDI-TOF mass spectra of PDIM and mycolic acids isolated from M. tuberculosis WT (B), opp mutant (opp KO) (C), and complemented strains (D), at different growth stages. OD, optical density 600 nm.
The opp KO strain shows an altered chronic phase of infection compared to WT
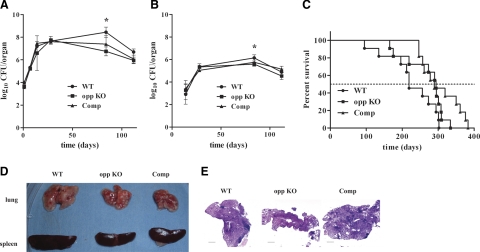
Virulence of the opp KO strain was evaluated using aerosol inoculation of BALB/c mice as a model. Previous studies have implicated the participation of Opp during infection (9, 10), and both PDIMs and Mycs are well-known mycobacterial virulence factors (28, 43). During the active phase (up to 1 mo postinfection), replication of the WT and opp KO strains was similar; however, on entry into chronic phase (12 wk postinfection), the opp KO mutant burden was significantly lower in both mouse lungs (P=0.004, Fig. 4A) and spleens (P=0.015, Fig. 4B) compared to the WT. However, introduction of the Opp locus into the opp KO mutant did not restore WT levels of replication (lungs, P=0.2; spleens, P=0.47, Fig. 4A, B). Persistence (defined here as the capacity to continue replication after 16 wk of infection) was not different between the WT parent, the opp KO (lungs P=0.6; spleens P 0.14), or the complemented KO (lungs P=0.09, spleens P=0.21). Nevertheless, we observed a delayed time-to-death trend for opp KO-infected animals compared to the WT, with median survival times of 295 and 220 d, respectively, that approached, but did not reach, statistical significance (P=0.076) (Fig. 4C). The complemented bacteria showed WT-like virulence (MST=289 d), although with statistical difference compared to it (P=0.011). Nevertheless, replication of the complemented strain was not statistically different to that of the opp KO mutant (P=0.13). Differences in granulomatous pathology were only minor at the organ level (Fig. 4D), and in the hematoxylin-eosin stained lung sections changes were not observed between the WT, KO, and complemented strains with the possible exception of wk 12, when inflammatory cell infiltrates were present in mice infected with the WT parent, but not in the lungs of mice infected with the mutant or complemented mutant (Fig. 4E). At wk 16, all lungs from mice infected with any of the strains had abundant inflammatory infiltrates (Fig. 4E).
Figure 4.
Lack of Opp affects the chronic phase of tuberculosis infection in mice. A, B) Bacterial CFUs (y axis) present in mouse lungs (A) and spleens (B) were log10 transformed and plotted against time postinfection (x axis). C) Survival kinetics of WT BALB/c mice infected via aerosol with 103 CFUs of either M. tuberculosis WT (circles), opp KO (squares), or the complemented opp KO strain (triangles). Surviving mice (y axis) are plotted throughout the time postinfection (x axis). D, E) Representative photographs of gross pathology (D) and histopathology (E) of bacterial infected tissues. Scale bars = 0.5 mm.
DISCUSSION
Mycobacteria belong to the same taxonomic order (Actinomycetales) as Streptomyces coelicolor and Corynebacterium diphtaeriae (44). Related species can serve as surrogate models from which metabolic or regulatory functions may be extrapolated, or new functions discovered from the analysis of orthologous genes. A compelling example of such an approach comes from recent studies focused on the WhiB protein family conserved in actinomycetes (45). A member of this family was shown to be required for intrinsic antibiotic resistance in both Mycobacterium and Streptomyces species (46).
In Streptomyces, Opp is thought to reimport a signaling molecule, an extracellular oligopeptide implicated in regulating development (19, 47). The absence of mutants in genes required to produce the signal may imply that this is a product of an essential process, or simply that the signal is the product of a small gene that is rarely hit during mutagenesis (48). Our attempts to identify a signaling peptide in Mtb-conditioned culture medium were unsuccessful, perhaps because of the lack of a distinctive phenotype for screening (e.g., sporulation in Bacillus or aerial hyphae formation in Streptomyces), although recently Ghosh et al. suggested sporulation occurs in Mycobacterium marinum and M. bovis var. BCG (49), thus raising the question whether further attempts to discover a putative signaling peptide should be conducted under the experimental conditions they used.
Phenotypic characterization of the opp KO mutant with the toxic tripeptide bialaphos showed that the current annotation of di- and oligopeptide transport systems in slow-growing mycobacteria needs to be modified. The demonstration that disruption of the Rv3666c-3663c locus results in bialaphos resistance shows that this locus encodes Mtb Opp rather than Rv1283c-1280c as previously believed (8). Consistent with this prediction is the demonstration that orthologues of the Mtb Rv3666c-3663c region in other genera also specify susceptibility to bialaphos. Like the Mtb Rv3665c-3663 mutant studied here (Fig. 1A), opp mutants of Bacillus (1, 18), Streptomyces (19, 20), and Listeria (21) were found to be resistant to bialaphos. This result also explains why Green et al. (8) were not able to show bialaphos resistance in an Rv1281c mutant of BCG. Instead, disruption of Rv1281c resulted in resistance to the toxic effects of glutathione (GSH) and the dipeptide l-cystenylglycine (8). Consistent with the idea that the Rv1283c-1280c locus encodes Dpp is the observation that bacterial degradation of GSH proceeds outside the plasma membrane via γ-glutamyl transpeptidase, leading to formation of γ-glutamyl residue and l-cystenylglycine. This in turn undergoes cleavage by dipeptidase to cystine and glycine, which are internalized into the cell (50). Mtb possesses one probable ggt gene (Rv2394, ggtB) whose predicted topology suggests outer surface activity (http:// www.doembi.ucla.edu/TB/PUBLIC/qs/qsearch.php?orf= Rv2394) but lacks a homologous gene for l-cysteinylglycine dipeptidase activity, thus likely explaining toxicity in WT cells following internalization of this dipeptide. Consequently, as observed by Green et al. (8), the Rv1283c-1280c mutant would not internalize l-cysteinylglycine, thus rendering the mutant resistant to this dipeptide.
Microarray expression profiling suggested that Mtb Opp, as is the case in other species, might modulate cell surface architecture by altering the expression of virulence-associated lipid and protein components (Fig. 2). Consistent with this idea were lipid analysis results showing clear differences between the WT and the opp KO mutant (Fig. 2). During early logarithmic growth phase, the opp KO strain accumulated more TAGs (Fig. 3A) than the WT, and this phenomenon reverted on complementation (Fig. 3A). TAG accumulation has been observed for Mtb present in sputa (51) and when bacteria enter a dormancy-like state in vitro (52). A comparison of members of the Opp regulon and genes induced by conditions likely encountered by Mtb during chronic infection (i.e., nutrient and/or oxygen deprivation) revealed that some overlap exists with transcriptional response to the latter: Rv0467 (icl), Rv2245 (kasA), Rv2246 (kasB), Rv2524 (fas), Rv2931 (ppsA), Rv2932 (ppsB), and Rv2933 (ppsC) are induced in response to oxygen deprivation and were modulated by Opp (Fig. 2 and Table 1) (53). Conversely, during nutrient deprivation by growth in PBS, fas, ppsA, ppsB, and ppsC were down-regulated (54). Perhaps metabolic perturbations occurring during hypoxic conditions resemble to some extent those occurring in the absence of Opp, and in turn affect expression of some genes required for virulence or chronic infection.
Nutrient deprivation occurring as a consequence of Opp absence seems unlikely, at least under the experimental conditions we tested. The WT and opp KO mutant strains grew to the same extent in vitro (Fig. 1) and in vivo, except during the chronic phase of infection (Fig. 4). The disruption of other Mtb genes have yielded mutants impaired in their capacity to sustain the chronic phase of infection in murine models. These include gene-encoding metabolic functions, like icl and mel2 (55, 56), and gene-encoding transporter functions, such as mce4 (57).
From this perspective, we regard Opp as having a broad biological role as a signaling component involved in modulating cell wall properties and interaction of the organism with surfaces. Examples supporting this view are the inability of Streptomyces opp mutants to form aerial hyphae, thus being referred to as “bald” (bld) (19), or the incapacity to remodel the cell envelope or modify intracellular phosphorelay cascades to form spores in Bacillus (18). Moreover, opp mutants of Streptococcus pneumoniae (58, 59) and Streptococcus agalactiae (60) have an altered capacity to attach to host cells. In support of the view that the Mtb opp KO mutant may have altered surfaces is the number of cell-surface exposed protein (HbhA, LpqH, PPE-family proteins) and lipid (PDIM and mycolic acids) virulence-associated genes that were differentially expressed between WT and opp KO mutant in Mtb (Fig. 2).
It could well be that signaling events triggered by small signaling peptides produced in response to changes in environmental factors, such as nutrient composition or availability, are complexly intertwined, hindering their dissection. Over a decade ago, Pope et al. (61) demonstrated that the differentiation defect of S. coelicolor opp mutants that caused the bald phenotype was dependent on the carbon source in the medium: defects evident during growth in a minimal medium containing glucose are partially rescued by using mannitol as a carbon source. Moreover, a decreased level of inosine 5′-monophosphate (IMP) dehydrogenase (directly involved in the synthesis of GTP) was observed in a S. coelicolor crp− mutant and proposed to partially explain the early and activated morphological differentiation of this strain (62). A decrease in GTP level is proposed to be a critical parameter to initiate morphological differentiation in Streptomyces (39). However, none of these studies tested the bldK (opp) mutant, thus maintaining an open question for this particular component: whether changing the carbon source or supplementing the medium with GTP could metabolically rescue the morphological differentiation defect.
MALDI-TOF analysis of the alkaline methanolysis products of extractable lipids showed that, using PDIM intensity peaks as a 100% reference, the relative intensities of peaks corresponding to mycolic acids varied in a growth-phase-dependent manner (Fig. 3B). The opp KO strain showed altered PDIM/Myc ratio (Fig. 3C), further suggesting that regulatory events programming cell wall content are affected in the mutant. The PDIM/Myc ratio returned to WT-like levels in the complemented strain (Fig. 3D). We found no significant differences at any time in cell wall-linked mycolates. Therefore, the observed variation in the PDIM/Myc ratio in lipids extracted from the opp KO mutant suggests that it is the PDIM component that changed.
Regarding the impact of Opp on virulence in other bacterial pathogens, a Listeria monocytogenes oppA mutant was attenuated for growth at low temperatures (5°C), affected for survival inside macrophages, and had a slightly delayed time-to-death phenotype in mouse infection (21). An S. pneumoniae opp mutant showed diminished binding to type II pneumocytes and endothelial cells in vitro (58) as well as reduced nasopharyngeal colonization in vivo (59). An S. agalactiae oppB mutant showed reduced adherence to epithelial cells (60), and a Streptococcus pyogenes oppA mutant showed altered expression of virulence factors and diminished virulence in a mouse model (63).
Both in vitro and in vivo studies have implicated the opp locus as relevant for mycobacterial infection. An M. bovis var. BCG mutant derived from a transposon insertion in Rv3664c showed a severely diminished capacity to survive in murine macrophages (9), whereas in Mtb H37Rv, Rv3663c was required for in vivo survival after intravenous infection of C57BL/6J mice (10). In human infected lungs, Rv3663c, Rv3664c, and Rv3665c were among genes induced in pericavitary lesions (where most bacilli reside within macrophages) compared to cells grown in vitro (64).
Based on these findings, and our observation of the altered cell wall content of the Mtb opp KO strain, including PDIM, a putative virulence determinant, we investigated the capacity of this mutant to cause disease in aerosol-infected BALB/c mice. In this model, Opp was required to maintain bacterial burden during the chronic, but not the persistent, phase of infection (Fig. 4A, B). Opp was also required to promote inflammatory cell infiltrates at chronic stage (Fig. 4E). Nevertheless, these defects faded once reaching a persistent phase of disease, resulting in a slightly (and close to statistically significant) decreased capacity to kill animals (Fig. 4C). Despite the fact that complementation of the opp KO mutant restored in vitro phenotypes, the complemented mutant did not achieve WT levels in the bacterial burden during the chronic phase of infection (Fig. 4A, B). This could be due to differences during in vivo expression of OppB-D, owing to its presence at the chromosomal attB site, having a different genomic context with respect to its WT location (65).
These results are seemingly in disagreement with Opp having an important role for in vivo survival. Possibly this can be explained by variations in replicative capacity, virulence, and pathology and how these factors are affected by bacterial genotype, mouse genotype, the infection model used, and the challenge dose (Table 2, 11, 12, 66,67,68,69,70,71,72,73,74). A striking in vivo virulence phenotype may not have been detected because cell wall changes in the PDIM/Myc ratio that were seen when the opp KO mutant was grown in vitro are not highly relevant in the model tested. In mice, other subtle cell-wall changes occuring to Mtb, such as oxygenation as well as cis- or trans-cyclopropanation of mycolic acids, dramatically affected infection outcome (75–77). Taken together, these observations reinforce the need to carefully consider how virulence is defined and the importance of animal model selection on the presence and magnitude of a mutant’s virulence phenotype.
TABLE 2.
Comparison of virulence of M. tuberculosis strains evaluated in mouse models
| M. tuberculosis strain | Mouse | Median survival time | Log10 CFU implanted | Route of infection | Reference |
|---|---|---|---|---|---|
| CDC1551 | C57BL/6 | 36 wk | 2 | Aerosol | 67 |
| CDC1551 | C3H/HeJ | 25 wk | 2 | Aerosol | 68 |
| CDC1551 | C3H/HeJ | 11 wk | 6 | Intravenous | 69 |
| CDC1551 | BALB/c | 23 wk | 6 | Intravenous | 70 |
| CDC1551 | C3H/HeJ | 7 wk | 6 | Intravenous | 5 |
| CDC1551 | C57BL/6 | No dead animals | 2 | Aerosol | 5 |
| CDC1551 | DBA/2 | 25 wk | 2 | Aerosol | 71 |
| CDC1551 | DBA/2 | 13 wk | 6 | Intravenous | 71 |
| CDC1551 | B6D2a | 38 wk | 2.4 | Aerosol | 72 |
| H37Rv | B6D2a | 26 wk | 2.2 | Aerosol | 72 |
| H37Rv | DBA/2 | 12 wk | 2 | Aerosol | 73 |
| H37Rv | BALB/c | 14 wk | 6 | Intravenous | 74 |
| H37Rv | BALB/c | 17 wk | 6 | Intravenous | 75 |
| H37Rv | C57BL/6 | 21 wk | 6 | Intravenous | 4 |
| H37Rv | BALB/c | No dead animals | 1.5 | Aerosol | P. Converse, personal communication |
| HN878 | B6D2a | 18 wk | 3 | Aerosol | 72 |
| 1254 | BALB/c | 31 wk | 3.6 | Aerosol | This work |
(C57BL/6×DBA/2)F1 (B6D2F1).
Acknowledgments
The support of NIH contract N01-AI30036 is gratefully acknowledged. Within this group, our especial appreciation to Dr. Gyanu Lamichhane, who designed animal infection experiments, Dr. Paul Converse for his advice and data analysis, and Scott Nolan for superior technical assistance. The authors thank Dr. Jeannette Barba for her assistance in exporting microarray data to GEO and ArrayExpress. M.A.F.V. was a recipient of the Stanford University School of Medicine Dean’s Fellowship.
References
- Solomon J M, Lazazzera B A, Grossman A D. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev. 1996;10:2014–2024. doi: 10.1101/gad.10.16.2014. [DOI] [PubMed] [Google Scholar]
- Lazazzera B A, Solomon J M, Grossman A D. An exported peptide functions intracellularly to contribute to cell density signaling in B. subtilis. Cell. 1997;89:917–925. doi: 10.1016/s0092-8674(00)80277-9. [DOI] [PubMed] [Google Scholar]
- Lazazzera B A. The intracellular function of extracellular signaling peptides. Peptides. 2001;22:1519–1527. doi: 10.1016/s0196-9781(01)00488-0. [DOI] [PubMed] [Google Scholar]
- Gominet M, Slamti L, Gilois N, Rose M, Lereclus D. Oligopeptide permease is required for expression of the Bacillus thuringiensis plcR regulon and for virulence. Mol Microbiol. 2001;40:963–975. doi: 10.1046/j.1365-2958.2001.02440.x. [DOI] [PubMed] [Google Scholar]
- Slamti L, Lereclus D. A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J. 2002;21:4550–4559. doi: 10.1093/emboj/cdf450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard B A, Podbielski A, Hedberg P J, Dunny G M. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc Natl Acad Sci U S A. 1996;93:260–264. doi: 10.1073/pnas.93.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M, Hoch J A. Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proc Natl Acad Sci U S A. 1996;93:1549–1553. doi: 10.1073/pnas.93.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R M, Seth A, Connell N D. A peptide permease mutant of Mycobacterium bovis BCG resistant to the toxic peptides glutathione and S-nitrosoglutathione. Infect Immun. 2000;68:429–436. doi: 10.1128/iai.68.2.429-436.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G R, Patel J, Robertson B D, Rae A, Young D B. Mycobacterial mutants with defective control of phagosomal acidification. PLoS Pathog. 2005;1:269–278. doi: 10.1371/journal.ppat.0010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti C M, Rubin E J. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn A J, Collins D M, Hondalus M K, Jacobs W R, Jr, Kawakami R P, Bloom B R. Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc Natl Acad Sci U S A. 2002;99:3147–3152. doi: 10.1073/pnas.052705399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal D, Schroeder B G, Tyagi S, Yoshimatsu T, Scott C, Ko C, Carpenter L, Mehrotra J, Manabe Y C, Fleischmann R D, Bishai W R. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc Natl Acad Sci U S A. 2002;99:8330–8335. doi: 10.1073/pnas.102055799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardarov S, Bardarov S, Jr, Pavelka M S, Jr, Sambandamurthy V, Larsen M, Tufariello J, Chan J, Hatfull G, Jacobs W R., Jr Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology. 2002;148:3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- Sassetti C M, Boyd D H, Rubin E J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Fleischmann R D, Alland D, Eisen J A, Carpenter L, White O, Peterson J, DeBoy R, Dodson R, Gwinn M, Haft D, Hickey E, Kolonay J F, Nelson W C, Umayam L A, Ermolaeva M, Salzberg S L, Delcher A, Utterback T, Weidman J, Khouri H, Gill J, Mikula A, Bishai W, Jacobs W R, Jr, Venter J C, Fraser C M. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J Bacteriol. 2002;184:5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, Berger S, Heinzelmann E, Muschko K, Welzel K, Wohlleben W. Biosynthetic gene cluster of the herbicide phosphinothricin tripeptide from Streptomyces viridochromogenes Tu494. Appl Environ Microbiol. 2004;70:7093–7102. doi: 10.1128/AEM.70.12.7093-7102.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M, Higgins C F, Pearce S R, Gallagher M P, Hoch J A. The oligopeptide transport system of Bacillus subtilis plays a role in the initiation of sporulation. Mol Microbiol. 1991;5:173–185. doi: 10.1111/j.1365-2958.1991.tb01838.x. [DOI] [PubMed] [Google Scholar]
- Nodwell J R, McGovern K, Losick R. An oligopeptide permease responsible for the import of an extracellular signal governing aerial mycelium formation in Streptomyces coelicolor. Mol Microbiol. 1996;22:881–893. doi: 10.1046/j.1365-2958.1996.01540.x. [DOI] [PubMed] [Google Scholar]
- Lorenzana L M, Perez-Redondo R, Santamarta I, Martin J F, Liras P. Two oligopeptide-permease-encoding genes in the clavulanic acid cluster of Streptomyces clavuligerus are essential for production of the beta-lactamase inhibitor. J Bacteriol. 2004;186:3431–3438. doi: 10.1128/JB.186.11.3431-3438.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borezee E, Pellegrini E, Berche P. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect Immun. 2000;68:7069–7077. doi: 10.1128/iai.68.12.7069-7077.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrakchi H, Bardou F, Lanéelle M A, Daffé M. A comprehensive overview of the mycolic acid biosynthesis. Daffé M, Reyrat J M, editors. Washington, DC: ASM Press; The Mycobacterial Cell Envelope. 2008:41–62. [Google Scholar]
- Barry C E, Crick D C, McNeil M R. Targeting the formation of the cell wall core of M. tuberculosis. Infect Disord Drug Targets. 2007;7:182–202. doi: 10.2174/187152607781001808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi O A, Arora P, Vats A, Ansari M Z, Tickoo R, Sridharan V, Mohanty D, Gokhale R S. Dissecting the mechanism and assembly of a complex virulence mycobacterial lipid. Mol Cell. 2005;17:631–643. doi: 10.1016/j.molcel.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Cox J S, Chen B, McNeil M, Jacobs W R., Jr Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- Camacho L R, Ensergueix D, Perez E, Gicquel B, Guilhot C. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol Microbiol. 1999;34:257–267. doi: 10.1046/j.1365-2958.1999.01593.x. [DOI] [PubMed] [Google Scholar]
- Guilhot C A D M. Polyketides and polyketide-containing glycolipids of M. tuberculosis: structure, biosynthesis and biological activities. Kaufmann S H E, Rubin E J, editors. Weinheim, Germany: Wiley-VCH; 2008:21–51. [Google Scholar]
- Jackson M, Stadthagen G, Gicquel B. Long-chain multiple methyl-branched fatty acid-containing lipids of Mycobacterium tuberculosis: biosynthesis, transport, regulation and biological activities. Tuberculosis (Edinb) 2007;87:78–86. doi: 10.1016/j.tube.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Dubey V S, Sirakova T D, Kolattukudy P E. Disruption of msl3 abolishes the synthesis of mycolipanoic and mycolipenic acids required for polyacyltrehalose synthesis in Mycobacterium tuberculosis H37Rv and causes cell aggregation. Mol Microbiol. 2002;45:1451–1459. doi: 10.1046/j.1365-2958.2002.03119.x. [DOI] [PubMed] [Google Scholar]
- Lamb J R, Rees A D, Bal V, Ikeda H, Wilkinson D, De Vries R R, Rothbard J B. Prediction and identification of an HLA-DR-restricted T cell determinant in the 19-kDa protein of Mycobacterium tuberculosis. Eur J Immunol. 1988;18:973–976. doi: 10.1002/eji.1830180623. [DOI] [PubMed] [Google Scholar]
- Ashbridge K R, Prestidge R L, Booth R J, Watson J D. The mapping of an antibody-binding region on the Mycobacterium tuberculosis 19 kilodalton antigen. J Immunol. 1990;144:3137–3142. [PubMed] [Google Scholar]
- Sorensen A L, Nagai S, Houen G, Andersen P, Andersen A B. Purification and characterization of a low-molecular-mass T-cell antigen secreted by Mycobacterium tuberculosis. Infect Immun. 1995;63:1710–1717. doi: 10.1128/iai.63.5.1710-1717.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet F X, Rasmussen P B, Rosenkrands I, Andersen P, Gicquel B. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10) Microbiology. 1998;144:3195–3203. doi: 10.1099/00221287-144-11-3195. [DOI] [PubMed] [Google Scholar]
- Pethe K, Alonso S, Biet F, Delogu G, Brennan M J, Locht C, Menozzi F D. The heparin-binding haemagglutinin of M. tuberculosis is required for extrapulmonary dissemination. Nature. 2001;412:190–194. doi: 10.1038/35084083. [DOI] [PubMed] [Google Scholar]
- Zvi A, Ariel N, Fulkerson J, Sadoff J C, Shafferman A. Whole genome identification of Mycobacterium tuberculosis vaccine candidates by comprehensive data mining and bioinformatic analyses. BMC Med Genomics. 2008;1:18. doi: 10.1186/1755-8794-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnappinger D, Ehrt S, Voskuil M I, Liu Y, Mangan J A, Monahan I M, Dolganov G, Efron B, Butcher P D, Nathan C, Schoolnik G K. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Elias E J, McKinney J D. Carbon metabolism of intracellular bacteria. Cell Microbiol. 2006;8:10–22. doi: 10.1111/j.1462-5822.2005.00648.x. [DOI] [PubMed] [Google Scholar]
- Ochi K, Kandala J C, Freese E. Initiation of Bacillus subtilis sporulation by the stringent response to partial amino acid deprivation. J Biol Chem. 1981;256:6866–6875. [PubMed] [Google Scholar]
- Ochi K. Metabolic initiation of differentiation and secondary metabolism by Streptomyces griseus: significance of the stringent response (ppGpp) and GTP content in relation to A factor. J Bacteriol. 1987;169:3608–3616. doi: 10.1128/jb.169.8.3608-3616.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englebrecht H L. Time course of purine nucleoside phosphorylase occurrence in sporulation of Bacillus cereus. J Bacteriol. 1972;111:33–36. doi: 10.1128/jb.111.1.33-36.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laval F, Laneelle M A, Deon C, Monsarrat B, Daffe M. Accurate molecular mass determination of mycolic acids by MALDI-TOF mass spectrometry. Anal Chem. 2001;73:4537–4544. doi: 10.1021/ac0105181. [DOI] [PubMed] [Google Scholar]
- Camacho L R, Constant P, Raynaud C, Laneelle M A, Triccas J A, Gicquel B, Daffe M, Guilhot C. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis: evidence that this lipid is involved in the cell wall permeability barrier. J Biol Chem. 2001;276:19845–19854. doi: 10.1074/jbc.M100662200. [DOI] [PubMed] [Google Scholar]
- Brennan P J. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2003;83:91–97. doi: 10.1016/s1472-9792(02)00089-6. [DOI] [PubMed] [Google Scholar]
- Bentley S D, Chater K F, Cerdeno-Tarraga A M, Challis G L, Thomson N R, James K D, Harris D E, Quail M A, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen C W, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang C H, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream M A, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell B G, Parkhill J, Hopwood D A. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald G F, Chater K F, van Sinderen D. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris R P, Nguyen L, Gatfield J, Visconti K, Nguyen K, Schnappinger D, Ehrt S, Liu Y, Heifets L, Pieters J, Schoolnik G, Thompson C J. Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2005;102:12200–12205. doi: 10.1073/pnas.0505446102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodwell J R, Losick R. Purification of an extracellular signaling molecule involved in production of aerial mycelium by Streptomyces coelicolor. J Bacteriol. 1998;180:1334–1337. doi: 10.1128/jb.180.5.1334-1337.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater K F, Horinouchi S. Signalling early developmental events in two highly diverged Streptomyces species. Mol Microbiol. 2003;48:9–15. doi: 10.1046/j.1365-2958.2003.03476.x. [DOI] [PubMed] [Google Scholar]
- Ghosh J, Larsson P, Singh B, Pettersson B M, Islam N M, Sarkar S N, Dasgupta S, Kirsebom L A. Sporulation in mycobacteria. Proc Natl Acad Sci U S A. 2009;106:10781–10786. doi: 10.1073/pnas.0904104106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova G V, Oktyabrsky O N. Glutathione in bacteria. Biochemistry (Mosc) 2005;70:1199–1211. doi: 10.1007/s10541-005-0248-3. [DOI] [PubMed] [Google Scholar]
- Garton N J, Waddell S J, Sherratt A L, Lee S M, Smith R J, Senner C, Hinds J, Rajakumar K, Adegbola R A, Besra G S, Butcher P D, Barer M R. Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. PLoS Med. 2008;5:e75. doi: 10.1371/journal.pmed.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J, Deb C, Dubey V S, Sirakova T D, Abomoelak B, Morbidoni H R, Kolattukudy P E. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J Bacteriol. 2004;186:5017–5030. doi: 10.1128/JB.186.15.5017-5030.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H D, Guinn K M, Harrell M I, Liao R, Voskuil M I, Tompa M, Schoolnik G K, Sherman D R. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol. 2003;48:833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts J C, Lukey P T, Robb L C, McAdam R A, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- McKinney J D, Honer zu Bentrup K, Munoz-Elias E J, Miczak A, Chen B, Chan W T, Swenson D, Sacchettini J C, Jacobs W R, Jr, Russell D G. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- Cirillo S L, Subbian S, Chen B, Weisbrod T R, Jacobs W R, Jr, Cirillo J D. Protection of Mycobacterium tuberculosis from reactive oxygen species conferred by the mel2 locus impacts persistence and dissemination. Infect Immun. 2009;77:2557–2567. doi: 10.1128/IAI.01481-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A K, Sassetti C M. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci U S A. 2008;105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cundell D R, Pearce B J, Sandros J, Naughton A M, Masure H R. Peptide permeases from Streptococcus pneumoniae affect adherence to eucaryotic cells. Infect Immun. 1995;63:2493–2498. doi: 10.1128/iai.63.7.2493-2498.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr A R, Adrian P V, Estevao S, de Groot R, Alloing G, Claverys J P, Mitchell T J, Hermans P W. The Ami-AliA/AliB permease of Streptococcus pneumoniae is involved in nasopharyngeal colonization but not in invasive disease. Infect Immun. 2004;72:3902–3906. doi: 10.1128/IAI.72.7.3902-3906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samen U, Gottschalk B, Eikmanns B J, Reinscheid D J. Relevance of peptide uptake systems to the physiology and virulence of Streptococcus agalactiae. J Bacteriol. 2004;186:1398–1408. doi: 10.1128/JB.186.5.1398-1408.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope M K, Green B D, Westpheling J. The bld mutants of Streptomyces coelicolor are defective in the regulation of carbon utilization, morphogenesis and cell-cell signalling. Mol Microbiol. 1996;19:747–756. doi: 10.1046/j.1365-2958.1996.414933.x. [DOI] [PubMed] [Google Scholar]
- Piette A, Derouaux A, Gerkens P, Noens E E, Mazzucchelli G, Vion S, Koerten H K, Titgemeyer F, De Pauw E, Leprince P, van Wezel G P, Galleni M, Rigali S. From dormant to germinating spores of Streptomyces coelicolor A3(2): new perspectives from the crp null mutant. J Proteome Res. 2005;4:1699–1708. doi: 10.1021/pr050155b. [DOI] [PubMed] [Google Scholar]
- Wang C H, Lin C Y, Luo Y H, Tsai P J, Lin Y S, Lin M T, Chuang W J, Liu C C, Wu J J. Effects of oligopeptide permease in group a streptococcal infection. Infect Immun. 2005;73:2881–2890. doi: 10.1128/IAI.73.5.2881-2890.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachman H, Strong M, Ulrichs T, Grode L, Schuchhardt J, Mollenkopf H, Kosmiadi G A, Eisenberg D, Kaufmann S H. Unique transcriptome signature of Mycobacterium tuberculosis in pulmonary tuberculosis. Infect Immun. 2006;74:1233–1242. doi: 10.1128/IAI.74.2.1233-1242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover C K, de la Cruz V F, Fuerst T R, Burlein J E, Benson L A, Bennett L T, Bansal G P, Young J F, Lee M H, Hatfull G F, Snapper S B, Barletta R G, Jacobs W R, Jr, Bloom B R. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- Manca C, Tsenova L, Barry C E, 3rd, Bergtold A, Freeman S, Haslett P A, Musser J M, Freedman V H, Kaplan G. Mycobacterium tuberculosis CDC1551 induces a more vigorous host response in vivo and in vitro, but is not more virulent than other clinical isolates. J Immunol. 1999;162:6740–6746. [PubMed] [Google Scholar]
- Calamita H, Ko C, Tyagi S, Yoshimatsu T, Morrison N E, Bishai W R. The Mycobacterium tuberculosis SigD sigma factor controls the expression of ribosome-associated gene products in stationary phase and is required for full virulence. Cell Microbiol. 2005;7:233–244. doi: 10.1111/j.1462-5822.2004.00454.x. [DOI] [PubMed] [Google Scholar]
- Agarwal N, Woolwine S C, Tyagi S, Bishai W R. Characterization of the Mycobacterium tuberculosis sigma factor SigM by assessment of virulence and identification of SigM-dependent genes. Infect Immun. 2007;75:452–461. doi: 10.1128/IAI.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando M, Yoshimatsu T, Ko C, Converse P J, Bishai W R. Deletion of Mycobacterium tuberculosis sigma factor E results in delayed time to death with bacterial persistence in the lungs of aerosol-infected mice. Infect Immun. 2003;71:7170–7172. doi: 10.1128/IAI.71.12.7170-7172.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Ruiz R E, Li Q, Silver R F, Bishai W R. Construction and characterization of a Mycobacterium tuberculosis mutant lacking the alternate sigma factor gene, sigF. Infect Immun. 2000;68:5575–5580. doi: 10.1128/iai.68.10.5575-5580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Converse P J, Ko C, Tyagi S, Morrison N E, Bishai W R. Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol Microbiol. 2004;52:25–38. doi: 10.1111/j.1365-2958.2003.03958.x. [DOI] [PubMed] [Google Scholar]
- Dainese E, Rodrigue S, Delogu G, Provvedi R, Laflamme L, Brzezinski R, Fadda G, Smith I, Gaudreau L, Palu G, Manganelli R. Posttranslational regulation of Mycobacterium tuberculosis extracytoplasmic-function sigma factor sigma L and roles in virulence and in global regulation of gene expression. Infect Immun. 2006;74:2457–2461. doi: 10.1128/IAI.74.4.2457-2461.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M Y, Raman S, Anaya M, Husson R N. The Mycobacterium tuberculosis extracytoplasmic-function sigma factor SigL regulates polyketide synthases and secreted or membrane proteins and is required for virulence. J Bacteriol. 2005;187:7062–7071. doi: 10.1128/JB.187.20.7062-7071.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S, Hazra R, Dascher C C, Husson R N. Transcription regulation by the Mycobacterium tuberculosis alternative sigma factor SigD and its role in virulence. J Bacteriol. 2004;186:6605–6616. doi: 10.1128/JB.186.19.6605-6616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau E, Chan J, Raynaud C, Mohan V P, Laneelle M A, Yu K, Quemard A, Smith I, Daffe M. Oxygenated mycolic acids are necessary for virulence of Mycobacterium tuberculosis in mice. Mol Microbiol. 2000;36:630–637. doi: 10.1046/j.1365-2958.2000.01882.x. [DOI] [PubMed] [Google Scholar]
- Glickman M S, Cox J S, Jacobs W R., Jr A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis. Mol Cell. 2000;5:717–727. doi: 10.1016/s1097-2765(00)80250-6. [DOI] [PubMed] [Google Scholar]
- Rao V, Gao F, Chen B, Jacobs W R, Jr, Glickman M S. Trans-cyclopropanation of mycolic acids on trehalose dimycolate suppresses Mycobacterium tuberculosis-induced inflammation and virulence. J Clin Invest. 2006;116:1660–1667. doi: 10.1172/JCI27335. [DOI] [PMC free article] [PubMed] [Google Scholar]