Abstract
Various types of the staphylococcal cassette chromosome mec (SCCmec) are known to confer methicillin resistance on the human pathogen Staphylococcus aureus. Such cassettes are not always stably maintained. The present studies were aimed at identifying the mechanism underlying the in vivo conversion of methicillin-resistant S. aureus (MRSA) to methicillin-susceptible S. aureus (MSSA) derivatives as encountered in two patients suffering from pneumonia and an umbilicus infection, respectively. All MRSA and MSSA isolates identified belong to multilocus sequence type (MLST) 398, have spa type t034, and are Panton-Valentine leukocidin positive. Sequencing of 27,616 nucleotides from the chromosomal SCCmec insertion site in orfX to the hsdR gene for a restriction enzyme revealed a type V (5C2&5) SCCmec. Sequence comparisons show that parts of the cassette are highly similar to sequences within SCCmec elements from coagulase-negative staphylococci, indicating a possible common origin. The cassette investigated contains ccrC-carrying units on either side of its class C2b mec gene complex. In vivo loss of the mec gene complex was caused by recombination between the recombinase genes ccrC1 allele 8 and ccrC1 allele 10. In vitro, the SCCmec was very stable, and low-frequency MRSA-to-MSSA conversion was only observed when MRSA isolates were cultivated at 41°C for prolonged periods of time. In this case also, loss of the mec complex was due to ccrC gene recombination. Interestingly, the MRSA and MSSA isolates studied displayed no detectable differences in competitive growth and virulence, suggesting that the presence of the intact type V (5C2&5) SCCmec has no negative bearing on staphylococcal fitness under the conditions used.
Staphylococcus aureus is a Gram-positive bacterium that can be part of the normal human microbiota as a colonizer of the mucosal membranes and skin. About 20% of the human population carries this commensal bacterium without any clinical symptoms (12). However, S. aureus has the potential to cause a wide range of infections, including wound infections, skin abscesses, pneumonia, bacteremia, meningitis, and toxic shock syndrome (12). In the early 1960s, only 2 years after the introduction of methicillin as a drug against S. aureus infections, hospital-acquired methicillin-resistant S. aureus (HA-MRSA) strains were first isolated (8). Since the 1990s, virulent community-acquired MRSA (CA-MRSA) strains, which are characterized by the presence of the toxin Panton-Valentine leukocidin (PVL), have been encountered in the community and health care (8).
Typing of numerous S. aureus isolates using pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), analysis of the variable repeat region of the gene for staphylococcal protein A (spa typing), and related approaches has revealed the existence of 11 major clonal lineages of S. aureus (11). In 2003, a new lineage of MRSA was identified as a colonizer of pigs and pig farmers in The Netherlands (8). Based on PFGE, these strains were classified as nontypeable MRSA, because their DNA cannot be digested with the restriction enzyme SmaI due to the presence of a novel DNA methylation activity (2). MLST showed that these veterinary strains belong to sequence type ST398, for which spa types t011, t034, and t108 are the most frequently identified.
Methicillin resistance is associated with the presence of a chromosomal mobile genetic element called the staphylococcal cassette chromosome mec (SCCmec). Eight main SCCmec types (I to VIII) have so far been identified, the sizes of which vary from ∼20 up to ∼50 kb (8, 20). The SCCmec classification is based on the composition of two gene complexes within the cassette, namely, the mec gene complex and the ccr gene complex (3, 8, 20). Common to all SCCmec elements is the presence of a mec gene complex that consists of the highly conserved mecA gene and intact or truncated sets of its regulatory genes, mecRI and mecI, and insertion sequences IS431 or IS1272 (22). The mecA gene encodes the membrane-associated penicillin binding protein 2a (PBP2a), which is involved in peptidoglycan synthesis. PBP2a confers resistance against all β-lactam antibiotics due to low-affinity binding, thereby overcoming the inhibition of native penicillin binding proteins as conferred by these antibiotics (6, 8). The mec gene complexes identified have been divided into three major classes that are defined by the sequences surrounding mecA, namely, class A (including IS431-mecA-mecR1-mecI), class B (including IS431-mecA-ΔmecR1::IS1272), and class C (including IS431-mecA-ΔmecR1::IS431) (8, 17, 23). In the class B and C mec gene complexes, the insertion sequences IS1272 and IS431 have integrated into the 3′ end of mecR1, ing in an inactive regulator protein. Furthermore, each SCCmec contains one or two ccr genes for recombinase(s) that play an important role in insertion and excision of the cassette from the genome (8, 17). At least nine different types of ccr gene complexes can be distinguished. Five SCCmec elements of S. aureus contain the recombinase genes ccrA and ccrB, two contain ccrA, ccrB, and ccrC, two contain one ccrC gene, and one contains two ccrC genes (8, 17, 20). SCCmec insertion and excision always occur at the same site, which is named attB. This site is located within the so-called orfX that seems to be strictly conserved in S. aureus genomes (21).
It has been reported before that SCCmec elements are intrinsically unstable when MRSA strains are cultivated in vitro. For example, Donnio et al. reported the loss of mecA due to a deletion within a type IV cassette (9). Furthermore, it was shown for three different MRSA isolates carrying a type II SCCmec that the in vitro selection for resistance to vancomycin coincided with a conversion of MRSA to methicillin-susceptible S. aureus (MSSA) (26). The mecA gene-containing region that was lost in each of the MSSA derivatives differed in length. In one of the MSSA strains studied, the complete SCCmec was lost due to site-specific chromosomal excision. In the other two MSSA strains, partial deletion of the respective SCCmecs appeared to involve the IS431 sequences, suggesting a role for IS431-mediated transposition/recombination in the observed mecA loss.
The present studies were aimed at identifying the mechanism for a likely case of in vivo MRSA-to-MSSA conversion. Isogenic MRSA and MSSA strains were isolated from both a Dutch mother and her adopted daughter from Anhwei, China. The mother was hospitalized with severe community-acquired staphylococcal pneumonia, and the daughter suffered from an S. aureus umbilicus phlegmon during the hospital admission of the mother. Here, we show that all MRSA and MSSA isolates from mother and daughter belong to ST398 and are nontypeable by PFGE using SmaI for restriction of genomic DNA. The ST398 MRSA isolates contain a new type of SCCmec which will be referred to as the type V (5C2&5) SCCmec from strain UMCG-M4. This cassette is related to the type V (5C2&5) SCCmec which was recently described for an S. aureus ST59 isolate (19, 31). Our results show that the MSSA isolates from mother and daughter have lost the mec gene complex but still contain remnants of the cassette. Loss of mecA was due to homologous recombination between two ccrC genes within the intact SCCmec type V (5C2&5). The MRSA ST398 strains identified are highly stable in vitro, which is consistent with the view that MRSA-to-MSSA conversion had occurred in vivo.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. MRSA and MSSA strains were grown overnight at 37°C or 41°C using 5% sheep blood agar plates (Media Products, Groningen, The Netherlands), tryptone soy agar (TSA) plates (Becton Dickinson), tryptone soy broth (TSB; Oxoid), MRSA-selective plates (bioMérieux), B medium (1% peptone [Difco], 0.5% yeast extract [Gibco-BRL], 0.5% NaCl, 0.1% K2HPO4, 0.1% glucose), or B plates. MRSA-selective plates were used to distinguish MRSA and MSSA strains after in vitro recombination experiments.
TABLE 1.
S. aureus strains used in this study
| Strain | Source | Type | Site of isolation | Phenotype |
|---|---|---|---|---|
| UMCG-M1 | Mother | CA-MRSA | Pleural fluid | Pcr Emr Oxar Tcs |
| UMCG-M2 | MSSA | Nose | Pcr Emr Tcs | |
| UMCG-M3 | CA-MRSA | Pus, nose | Pcr Emr Oxar Tcs | |
| UMCG-M4 | CA-MRSA | Pus, furuncle, nose | Pcr Emr Oxar Tcs | |
| UMCG-D1 | Daughter | MSSA | Pus, furuncle | Pcr Emr Tcs |
| UMCG-D2 | MSSA | Pus, furuncle | Pcr Emr Tcs | |
| UMCG-D3 | MSSA | Umbilicus phlegmon | Pcr Emr Tcs | |
| UMCG-D4 | CA-MRSA | Rectum swab | Pcr Emr Oxar Tcs |
Pcr, penicillin resistant; Emr, erythromycin resistant; Oxar, oxacillin resistant; Tcs, tetracycline sensitive.
Antibiotic susceptibility testing.
Antibiotic susceptibility was determined by disk diffusion, as recommended by the European Committee on Antimicrobial Susceptibility Testing (http://www.eucast.org/), in accordance with the instructions of the disk supplier (Neo-Sensitabs; Rosco Diagnostica A/S, Taastrup, Denmark). The antibiotics tested included erythromycin, penicillin, tetracycline, and oxacillin (Oxoid). The following reference clinical breakpoints were used to classify strains as susceptible, intermediate, or resistant: erythromycin susceptible, ≥28 mm, intermediate, 23 to 27 mm, and resistant, <23 mm; penicillin susceptible, ≥28 mm, and resistant, <28 mm; tetracycline susceptible, ≥30 mm, intermediate, 28 to 29 mm, and resistant, <28 mm; and oxacillin susceptible, ≥ 20 mm, and resistant, <20 mm.
Growth analyses of MSSA and MRSA isolates.
Overnight cultures were diluted to an optical density at 600 nm (OD600) of 0.05 in B medium and incubated at 37°C. Erlenmeyer flasks (100 ml) were shaken at 200 rpm. Aliquots were removed at the time points indicated below, and bacterial growth was assessed by OD600 readings. Competitive in vitro growth of strains MSSA UMCG-M2 and MRSA UMCG-M4 was assessed by inoculating 107 CFU of each strain from an overnight culture into one 100-ml flask containing B broth (15). The strains were cultivated for six days in the absence of antibiotics. Every 24 h, the mixed culture was diluted from 10−2 to 10−8 and plated on B agar plates without antibiotic and on agar plates containing 4 μg/ml methicillin. The number of CFU of strain MRSA UMCG-M4 was determined by counting the number of bacteria on the methicillin-containing plates. The number of CFU of the MSSA UMCG-M2 strain was determined by determining the difference in the numbers of colonies on nonselective and methicillin-containing plates.
PFGE.
The preparation of genomic DNA and separation by pulsed-field gel electrophoresis (PFGE) were performed as described by Stam-Bolink et al. (30). Agarose plugs containing genomic DNA were prepared from overnight cultures and digested with the restriction enzymes SmaI (Roche) or EheI (Fermentas) at 25°C or 37°C, respectively. Each plug was placed into a well of 0.8% agarose gel, and the cleaved genomic DNA was subsequently separated by PFGE.
DNA isolation, PCR, MLST, and spa typing.
Genomic DNA was isolated with a Genelute bacterial genomic DNA kit (Sigma) according to the manufacturer's protocol with the following modifications. Prelysis was performed using lysostaphin (5 U/μl) (Ambi Products) and lysozyme (Sigma) (10 mg/ml) in the kit's solution A for 20 min. PCRs were performed with a DNA Engine Tetrad (MJ Research) or a Bio-Rad C1000 thermal cycler. Primers used in this study were obtained from Eurogentec (see Table S1 in the supplemental material). The amplification reaction mix contained 0.625 units Super Taq (HT Biotechnology Ltd.), 2 μl template DNA, 200 μM each deoxynucleoside triphosphate (Roche), 5 μl of 10× reaction buffer, 0.25 μM each primer, and 1.25 mM MgCl2 in a final volume of 50 μl. The temperatures and times that were used for the different PCRs are indicated in Table S1 in the supplemental material. Fragments A, B, C, and D (see Fig. 2) were obtained by long-range PCR with Extensor high-fidelity PCR master mix (Thermo Scientific) according to the instructions of the supplier, with the following modifications. For the primer combinations AR plus IS431-R (fragment A) and ccr-γF plus hsdR-R (fragment D), annealing was performed for 30 s at 54°C and the extension reaction was performed for 8 min at 68°C. For the combinations Font-1 plus ccr-γF (fragment B) and Font-2 plus ccr-γR (fragment C), annealing was performed at 50°C for 30 s and extension at 68°C for 8 min. Purification of PCR products was done using a high pure PCR purification kit from Roche. PCR products were visualized using a 1% agarose gel. Multilocus sequence typing (MLST) was performed according to the protocol described by Enright et al. (10). The sequences obtained for each locus were submitted to the Internet database (www.mlst.net), and the resulting allelic profiles were assigned to a sequence type (ST). Spa typing was performed using the primer combinations (1) indicated in Table S1 in the supplemental material. The resulting sequences were analyzed using the software available at http://spaserver.ridom.de/.
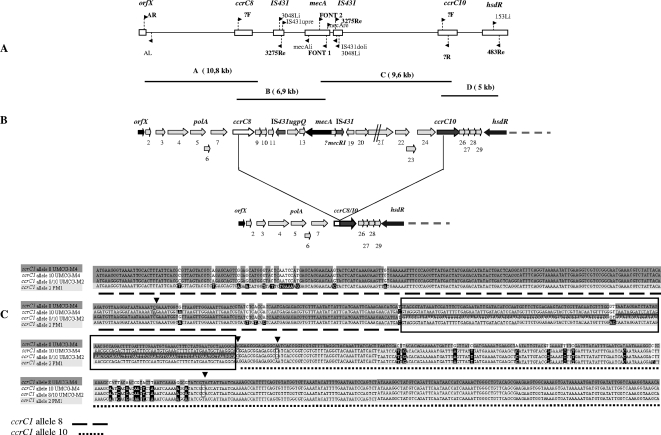
FIG. 2.
Mapping of mec and ccr gene complexes in the type V (5C2&5) SCCmec from strain UMCG-M4. (A) The location and orientation of primers used for amplification of the type V (5C2&5) SCCmec from UMCG-M4 fragments A, B, C, and D that were used for sequencing are indicated (see Table S1 in the supplemental material for primer sequences). (B) Schematic representation of the left side of the intact SCCmec type V (5C2&5) in strain UMCG-M4 and the region within this cassette that is absent from MSSA isolates. Gene names and numbers are indicated above the genes. The symbol “//” indicates stop codons or frameshift mutations in the putative ORFs. The recombination between ccrC1 allele 8 and ccrC1 allele 10 leading to MRSA-to-MSSA conversion is indicated. (C) Comparison of nucleotide sequences of ccrC genes from the type V (5C2&5) SCCmec of strain UMCG-M4 (ccrC1 allele 8 and ccrC1 allele 10), the recombined ccrC1 alleles 8/10 derived from the SCC element of strain UMCG-M2, and ccrC1 allele 2 from the type V (5C2&5) SCCmec of S. aureus PM1. Identical nucleotides in the depicted ccrC genes are marked with shades of gray (light and dark); nucleotides unique for ccrC1 allele 2 are shown with black boxes and white letters; nucleotides unique for ccrC1 allele 10 are shown in black frames with arrowheads; and identical nucleotides in ccrC1 allele 8, ccrC1 allele 10, and the recombinant ccrC1 alleles 8/10 are indicated with dark gray shading and white letters. The 142-bp region of the recombination sites in ccrC1 allele 8 and ccrC1 allele 10 is boxed.
Nucleotide sequence determination and analysis.
Sequencing was done by primer walking (ServiceXS, Leiden, The Netherlands). The sequences were compared to the nucleotide sequence database from the National Center for Biotechnology Information (NCBI) using the BLAST tool (http://www.ncbi.nlm.nih.gov/sites/entrez). Multiple sequence alignments were performed using ClustalW (http://align.genome.jp) software (www.ebi.ac.uk/clustalw). Signal peptides were predicted using the signalP algorithm (http://www.cbs.dtu.dk/services/SignalP/).
Southern hybridization.
Chromosomal DNA of all isolates was digested overnight with restriction enzyme HindIII or EcoRI at 37°C and separated on an 0.8% agarose gel. DNA was transferred to a membrane (Qiabrane) by diffusion blot. Probe hybridization and signal detection were performed using an Amersham ECL system according to the instructions of the supplier. The probe for hybridization was PCR amplified with primers AR and IS431-3275Re using chromosomal template DNA from MRSA strain UMCG-M4.
Screening for in vitro recombination.
Single colonies (strains UMCG-M4, UMCG-D4, UMCG-M2, and UMCG-D3) (Table 1) were picked from blood agar plates and used to inoculate 5 ml of fresh TSB medium. Upon overnight growth at 37°C, 5 μl of the culture was used to inoculate 5 ml of fresh medium, which was incubated overnight at 41°C. From the initial overnight cultures, 10−6 dilutions were plated on blood agar plates and on TSA plates, which were incubated overnight at 37°C. Single colonies were then transferred to MRSA-selective plates. Passage to fresh TSB medium, growth at 41°C, and plating were repeated every day for a period of 22 days.
Competitive virulence analysis.
All animal studies were approved by the Animal Care and Experimentation Committee of the district government of Lower Franconia, Germany, and conformed to University of Würzburg guidelines. Female BALB/c mice (16 to 18 g) purchased from Charles River, Sulzfeld, Germany, were housed in polypropylene cages and received food and water ad libitum. The S. aureus MSSA UMCG-M2 and MRSA UMCG-M4 strains were cultured for 18 h in B medium, washed 3 times with sterile 0.9% NaCl, and suspended in sterile 0.9% NaCl to 108 CFU/100 μl. To control viable cell counts, appropriate dilutions were plated on B agar. Mice were inoculated with 100 μl of the S. aureus strains MSSA UMCG-M2 and MRSA UMCG-M4, each via the tail vein. The experiments were performed with 6 mice. All mice were sacrificed by 120 h, and the kidneys were aseptically removed. The kidneys were homogenized using a Dispomix Drive device (Bio-Budget Technologies, Krefeld, Germany), and appropriate dilutions of the homogenates in 0.9% NaCl were plated on B agar plates with or without 4 μg/ml methicillin, respectively, to determine the bacterial load of the organs. Numbers of CFU for the UMCG-M2 and UMCG-M4 strains were calculated as indicated above. Statistical significance was tested using the Wilcoxon rank test.
Determination of the spontaneous mutation rate.
The mutation rate per cell and generation was calculated as described previously (13, 24, 27). Briefly, a single bacterial colony was diluted in 45 ml of phosphate-buffered saline. Amounts of 100 μl of this suspension were plated on agar plates to determine the inoculum size, and another 100-μl aliquot was used to inoculate 50 ml of B broth, which was grown at 37°C to an OD600 of 1.3 to 1.5. From this bacterial culture, 100-μl aliquots and appropriate dilutions were spread on B agar plates containing 10 μg of rifampin/ml. CFU were counted after 24 h of incubation at 37°C. The frequency of mutation (P) to rifampin resistance per cell and generation was calculated using the formula 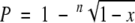 , where n is the number of generations and x is the number of rifampin-resistant colonies/total number of plated colonies. The number of generations (n) was determined using the equation
, where n is the number of generations and x is the number of rifampin-resistant colonies/total number of plated colonies. The number of generations (n) was determined using the equation 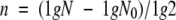 , where g is generation, N is the total number of bacterial cells in the culture, and N0 is the number of bacterial cells in the inoculum.
, where g is generation, N is the total number of bacterial cells in the culture, and N0 is the number of bacterial cells in the inoculum.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been deposited in the DDBJ/EMBL/GenBank database under accession no. GQ902038.
RESULTS
The MRSA isolates identified belong to the ST398 sequence type and contain a class C mec gene complex.
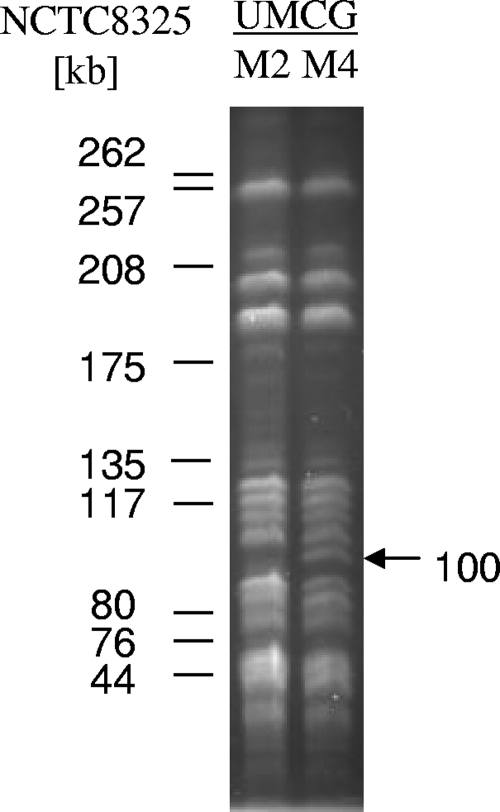
To test the relatedness of the MRSA and MSSA isolates from mother and daughter, several approaches were used. MLST analysis and spa typing showed that all isolates belong to the ST398 sequence type and spa type t034, which are associated with a veterinary origin in The Netherlands (33, 36). None of the isolates was typeable by PFGE using SmaI digestion. PFGE upon EheI digestion showed that the profiles from the MRSA isolates contained a fragment of ∼100 kb which was absent from the equivalent profiles of the MSSA isolates (Fig. 1). Antibiotic resistance profiling showed that all MRSA and MSSA isolates were resistant to penicillin and erythromycin (Table 1). An interesting finding was that all isolates were sensitive to tetracycline and tested positive for pvl in a PCR with specific primers (see Table S1 in the supplemental material). This showed that the isolates from mother and daughter differ from the Dutch pig-borne ST398 isolates, nearly all of which have so far tested tetracycline resistant and pvl negative (only one pvl-positive isolate has been reported so far) (33, 36).
FIG. 1.
PFGE comparison of chromosomal DNA isolated from MRSA (UMCG-M4) and MSSA (UMCG-M2) isolates. Chromosomal DNA was cleaved with restriction endonuclease EheI, and the fragments were separated by PFGE. The marker DNA fragments were obtained by restriction of chromosomal DNA of S. aureus strain NCTC8325 with EheI. The unique 100-kb EheI fragment observed for the MRSA UMCG-M4 isolate is indicated by an arrow.
As shown by PCR of genomic DNA, the MRSA isolates from mother and daughter (UMCG-D4, UMCG-M1, UMCG-M3, and UMCG-M4) contained mecA and IS431, while mecI appeared to be absent. As expected, mecA and IS431 could not be detected in the MSSA isolates. No products were obtained for ccr types AB, AB1, AB2, and AB3 or SCCmec IVa, -b, or -c. Importantly, two specific fragments of approximately 2 kb and 700 bp, respectively, were amplified with primers corresponding to IS431 and mecA, indicating that the SCCmec element in the genome of the MRSA strains contains a class C mec gene complex with the composition IS431-mecA-IS431 (Fig. 2A). While the mecA gene was absent from the MSSA isolates, these isolates carried the SCC J regions close to the orfX, ccrC, and hsdR genes, indicating that the MRSA-to-MSSA conversion had occurred through a deletion within the SCCmec being investigated.
The SCCmec from strain UMCG-M4 resembles a type V (5C2&5) SCCmec.
To elucidate the structure of the SCCmec from the ST398 MRSA and MSSA strains studied here, multiple PCRs were performed using different primer combinations for the identified genes (see Table S1 in the supplemental material). Four overlapping PCR fragments, named A to D, of ∼10.8, ∼6.9, ∼9.6, and ∼5 kb, respectively, which covered the area of interest were obtained for all MRSA isolates (Fig. 2A). For the MSSA strains, only fragment D was obtained (data not shown).
The nucleotide sequences of the PCR fragments of the SCCmec from strain UMCG-M4 were determined. The aligned sequence of 27,616 nucleotides contains 30 complete open reading frames (ORFs) (Fig. 2B). BLAST searches revealed that most of these ORFs code for unknown proteins (see Table S2 in the supplemental material). Sequencing of the orfX insertion point of SCCmec UMCG-M4 revealed the presence of the ISS consensus sequence (21), and in fact, the same insertion point was observed for the mecA-less SCC element of the MSSA isolate UMCG-M2. The sequences upstream and downstream of ISS were identical to the equivalent regions of the type V (5C2&5) SCCmec (previously indicated as type T) from the CA-MRSA strain PM1 (NCBI accession number AB353125) and SCCmecZH47 from the S. aureus strain ZH47 (NCBI accession number AM292304) (18). The similarity with the SCCmecZH47 continued to the left repeat of the IS431 element that is inserted into the mecR1 gene at nucleotide 14836 (Fig. 3). Thereafter, sequence similarity to part of the SCC region in the genome of Staphylococcus haemolyticus JCSC1435 (32) was observed, starting from nucleotide 14837 and ending in hsdR. A region of high similarity with the type V (5C2) SCCmec from strain JCSC3624 (WIS) (22) starts at position 11042 on the direct repeats located after the first IS431 element and continues to hsdR. The second IS431 transposase gene of the mec gene complex in the UMCG-M4 SCCmec contains two nucleotide mutations resulting in a truncated transposase. Identical mutations were found in the mec gene complexes of S. aureus strain PM1 (31) and S. haemolyticus JCSC1435 (32).
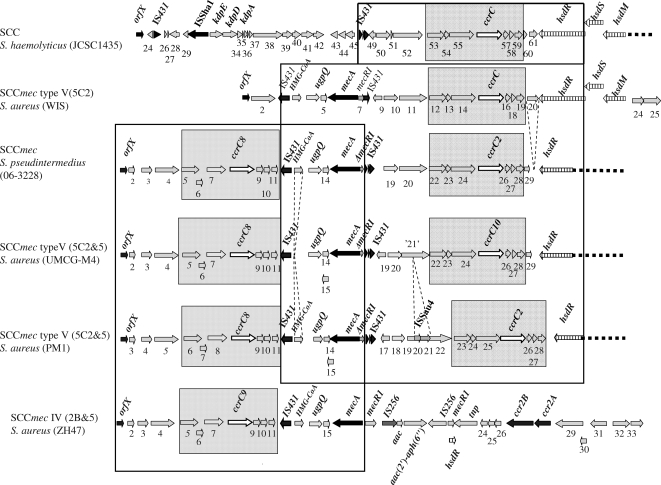
FIG. 3.
Comparison of the type V (5C2&5) SCCmec from strain UMCG-M4 with closely related SCCmec elements. The sequence determined for the type V (5C2&5) SCCmec from strain UMCG-M4 was compared with the following SCCmec sequences deposited in GenBank (accession numbers are indicated in parentheses): S. haemolyticus JCSC1435 (AP006716); S. aureus WIS (AB121219); S. pseudintermedius 06-3228 (FJ544922); S. aureus PM1 (AB353125); and S. aureus ZH47 (AM292304). Dark gray boxes indicate ccrC-carrying units in the SCCmec elements. Large frames indicate regions of high similarity between the type V (5C2&5) SCCmec from strain UMCG-M4 and other cassettes. Dotted lines indicate deletions of some ORFs. Genes are indicated by arrows. Black arrows indicate orfX, IS431, mecA, ccrA, and ccrB; white arrows indicate ccrCs; and gray arrows indicate hypothetical genes; ORF ‘21’ contains a stop codon. The hsdRSM restriction-modification system is indicated by white arrows with vertical lines. Names of functionally annotated genes are indicated above the respective arrows, and hypothetical genes are numbered.
The highest overall sequence similarities were observed with SCCmec sequences from Staphylococcus pseudintermedius strain 06-3228 (NCBI accession number FJ544922) and with the SCCmec type V (5C2&5) from the CA-MRSA S. aureus strain PM1 (19, 31). For this reason, we classified the SCCmec from strain UMCG-M4 as a type V (5C2&5) cassette. The SCCmec of UMCG-M4 lacks a region of 1,041 bp which is located between the left repeat of IS431 (nucleotides 11027/11042) and two direct repeats of 28 bp found in the SCCmec elements of strain PM1 and S. pseudintermedius (Fig. 3). This missing sequence encodes a putative hydroxymethylglutaryl coenzyme A (HMG-CoA) synthase. A second region missing from SCCmec UMCG-M4 is a region of 1,274 bp in the SCCmec of strain PM1 that encodes the ISSau4-like transposase (DQ680163.1|) (Fig. 3). This 1,274-bp region is flanked by inverted repeats (ACTGACCCC). In the SCCmec of strain PM1, the ORF for the transposase is interrupted, resulting in two ORFs (hypothetical genes numbered 20 and 21 [orf20 and orf21] in Fig. 3). orf21 of SCCmec of strain UMCG-M4 contains a frameshift in comparison to the corresponding genes in S. haemolyticus JCSC1435 and S. aureus strain JCSC3624 (WIS). Upstream of hsdR in strains WIS and S. haemolyticus JCSC1435, an additional gene is present (orf20 [AB121219.1|] and SH0061, respectively) (Fig. 3). This gene encodes a protein of 188 amino acids with a predicted signal peptide, suggesting that it is secreted. In both cases, the ORF is flanked on either side by the repeated sequence AGAGCATCCTTCACTTTTATGGTGAGGGATGCTCTTTTAATTTA that contains an internal inverted repeat (indicated by underlining). In the SCCmec type V (5C2&5) of strain UMCG-M4, the additional ORF is absent and the repeated sequence is present only once, directly upstream of the stop codon of hsdR.
Two regions running from nucleotides 349 to 3708 and 15854 to 19506 of the SCCmec of UMCG-M4 have a GC content of ∼27%. The sequence between these regions has a GC content of 32.6%, and the region downstream of the nucleotide at position 19506 has an average GC content of 33.7%. A similar distribution of the GC content was observed for the SCCmec sequences of S. aureus PM1 and S. pseudintermedius. This suggests that type V (5C2&5) cassettes are composed of units from different genetic origins, which is consistent with our observation that the SCCmec type V (5C2&5) from UMCG-M4 and other type V (5C2&5)(-like) SCCmec elements contain two regions of high similarity with type III and type V (5C2) SCCmec elements, respectively (Fig. 3).
Sequence conservation within the ccrC-carrying units.
Higuchi et al. indicated that the SCCmec of S. aureus strain PM1 contains two so-called ccrC-carrying units (19). Such units are also present in SCCmecs of S. pseudintermedius strain 06-3228 and UMCG-M4 (running from orf5 to orf11 and orf22 to orf28). BLAST searches showed that similar units are present only once in SCCmec types III, V (5C2), and VII (5C1), SCCmecZH47, SCCmercury, and the non-mecA SCC element SCCcap from different staphylococci. ClustalW comparisons of all these ccrC-carrying units showed that the nucleotide sequences homologous to orf5 and orf22, as well as ccrC1 allele 8 and ccrC1 allele 10, are very well conserved among all units, while the remaining sequences differ substantially. Dot plot analysis (http://www.vivo.colostate.edu/molkit/dnadot/) showed that these conserved nucleotide sequences in the type V (5C2&5) SCCmec from UMCG-M4 lead to large repeated regions running from nucleotide 3790 to 8789 and nucleotide 19404 to 24403. Similar repeats overlapping with the same genes were found in the nucleotide sequences of the SCCmec elements of S. aureus strain PM1 and S. pseudintermedius. Furthermore, the region from orf5 to ccrC1 allele 8 showed more than 97% sequence identity with the first ccrC-carrying units in the SCCmecs of S. aureus PM1 (19, 31) and S. pseudintermedius, while the region from orf22 to ccrC1 allele 10 showed only 91% sequence identity to the second ccrC-carrying units of those cassettes. This may suggest a higher level of conservation of the first ccrC-carrying unit. Most nucleotide differences were found in the regions encoding orf23 and orf24 of the UMCG-M4 SCCmec and the 3′ end of ccrC1 allele 10. Comparison of the nucleotide sequence of the primer ccr-γR with that of ccrC1 allele 8 showed that there is a mismatch in the 3′ end of ccr-γR, which contains a G instead of an A. This explained why no PCR products were obtained when the primers ccr-γR (corresponding to ccrC1 allele 8) and AR (corresponding to orfX) were used. Notably, ccrC1 allele 10 in the type V (5C2&5) SCCmec from UMCG-M4 represents a new allele type of the ccrC gene that differs from the known ccrC alleles.
Loss of the mec gene complex due to recombination between two ccrC genes.
Southern hybridization experiments for all eight isolates (MRSA/MSSA) showed the presence of two ccrC-carrying units in the MRSA isolates, while only one ccrC-carrying unit was detectable in the MSSA strains (data not shown). Since the MSSA isolates were known to have lost at least mecA from the mec gene complex, the primers up-ccrC8 and down-ccrC10 were used for PCR to determine the deletion point within the SCC element of strain UMCG-M2. This yielded a fragment of 5,047 bp, which was sequenced. Comparison with the DNA sequence of the complete SCCmec from UMCG-M4 showed that the 5,047-bp fragment lacked a stretch of 15,753 bp encoding all genes located between ccrC1 allele 8 and ccrC1 allele 10. Alignment of the nucleotide sequences of the ccrC genes present in the MRSA and MSSA strains studied (Fig. 2C) showed that the ccrC gene of the MSSA strains (referred to as ccrC1 allele 8/10) has resulted from a recombination within a region of 142 bp in the 3′ end of ccrC1 allele 8 and the 5′ end of ccrC1 allele 10 (Fig. 2B and C).
In vitro loss of the mec gene complex from the type V (5C2&5) SCCmec of strain UMCG-M4 can be induced by heat stress.
No MRSA-to-MSSA conversion was observed for the MRSA isolates from mother and daughter when these strains were cultivated in vitro at 37°C. This suggested that the MRSA isolates are very stable in vitro, at least with respect to maintenance of the mec gene complex. Mutator phenotypes are supposed to have adaptive advantages and play a role in the pathogenesis of bacterial infections, as well as in the development of antibiotic resistance (14). It has been shown that defects in mismatch repair systems which result in an elevated mutation rate contribute to the development of vancomycin resistance in S. aureus (29). In contrast, the S. aureus mutator phenotype was associated with a decreased bacterial fitness in vivo and did not confer a significant advantage in the acquisition of antibiotic resistance in a model of chronic bone infection (5). Therefore, we wanted to investigate whether or not a mutator phenotype was detectable among the MRSA UMCG-M4 and MSSA UMCG-M2 isolates and to elucidate possible differences in the mutation rates. For this purpose, we determined the spontaneous mutation rate in the rpoB gene, which confers resistance to rifampin (27). We have found that both strains had a spontaneous mutation frequency of 1 × 10−8. No difference with respect to the mutation rate was detectable between the two strains. Our data suggest that the S. aureus strains investigated in this study do not differ in the accumulation of point mutations under the experimental conditions applied.
To test whether MSSA isolates would have a competitive growth advantage over the corresponding MRSA isolates when cultivated in vitro, the MRSA UMCG-M4 and MSSA UMCG-M2 isolates were grown in Erlenmeyer flasks in B medium at 37°C. The results of comparative growth analyses showed that there was no detectable difference in the growth rates of the MRSA and MSSA isolates. Moreover, mixing of the isolates and subsequent cultivation in B medium for six days at 37°C revealed that the ratio between the isolates in the culture remained constant (data not shown). This demonstrates that the SCCmec being investigated is very stably maintained in the MRSA strains after their isolation from the patients.
To investigate whether MRSA-to-MSSA conversion could be observed under “stressful” in vitro growth conditions, single colonies of the MRSA strain UMCG-M4 were used to inoculate TSB standing cultures, which were incubated at 41°C. This temperature is known to induce homologous recombination in bacteria like S. aureus (25). After 3 weeks of daily transfers into fresh TSB and continued growth at 41°C, cells were plated on blood agar. The resulting colonies were then transferred to MRSA-selective plates. This screening resulted in the identification of four MSSA derivatives of MRSA strain UMCG-M4. PCR analysis with primers up-ccrC8 and down-ccrC10 yielded fragments of 5.1 kb for all four MSSA strains obtained in vitro, as was observed for the clinical MSSA isolates (data not shown). This showed that all four in vitro MSSA isolates had lost the mec gene complex by recombination between the ccrC-carrying units.
MRSA-to-MSSA conversion in the ST398 strains studied does not affect virulence.
To test whether MRSA-to-MSSA conversion might have affected the virulence of our isolates, a mouse kidney abscess model was used for infection experiments. The results revealed no significant difference in the virulence of the MRSA strain UMCG-M4 and the MSSA strain UMCG-M2 (data not shown). The mean bacterial load for MSSA strain UMCG-M2 was 6.2 × 106 CFU/organ, compared to 4.6 × 106 CFU/organ for MRSA strain UMCG-M4. These results suggest that the loss of methicillin resistance in the MSSA UMCG-M2 isolate had no impact on the competitive virulence of this strain compared to that of the MRSA UMCG-M4 isolate, which would explain, at least partially, why both MRSA and MSSA variants can coexist in patients.
DISCUSSION
In the present study, we document the in vivo conversion of MRSA to MSSA during “community-acquired” infections in which a mother and her adopted daughter suffered from pneumonia and an umbilicus phlegmon, respectively. The MRSA strains contain a type V (5C2&5)-like SCCmec from which the mec complex can be lost in vivo through recombination between two ccrC genes, revealing a novel mechanism for MRSA-to-MSSA conversion.
S. aureus ST398 strains are usually associated with a veterinary origin. Since it was first identified in isolates from pigs in The Netherlands in 2003, the S. aureus ST398 lineage has also been isolated from cattle, chickens, cats, dogs, and humans in Europe and China (16, 39, 42). In the case of the present study, there is no evidence for a direct transmission of the ST398 strain from animals to mother and/or daughter. In the literature, there is precedence for human transmission of ST398 strains (37, 41). Data from a recent outbreak with a nontypeable MRSA ST398 in The Netherlands (41) and from two ST398-related infections in Sweden (38) suggest that strains of this type can, in fact, be transferred quite readily between humans. Alternatively, ST398 strains may be transmitted through the food chain. In 2007, van Loo et al. reported that the pig- and cattle farm-related ST398 strain was detectable in Dutch meat products (35). Notably, the Dutch animal-related ST398 S. aureus strains are resistant to tetracycline, while the strains investigated in our studies are sensitive to this antibiotic (40). The adopted daughter was born in the Chinese province Anzhou, which is adjacent to the province Zhejiang where patients with pvl-positive ST398 MRSA have been identified. In contrast to the Chinese isolates, the vast majority of ST398 MRSA isolates in The Netherlands are pvl negative (42). These data suggest that the S. aureus ST398 isolates studied do not originate from The Netherlands but, possibly, from China.
Recently, the isolation of ST398 MRSA and MSSA strains belonging to spa types t011, t108, and t034 was reported (33, 39). MSSA ST398 spa t034 strains were found to be rare among healthy individuals, whereas relatively high numbers were derived from bacteremia patients. This led to the suggestion that MSSA ST398 spa t034 strains might be more virulent and compete for colonization space with the corresponding MRSA strains (4). The isogenic ST398 MRSA and MSSA isolates in the present study showed rates of growth in vitro and virulence properties in a mouse infection model that were indistinguishable. This implies that the isolated ST398 MSSA strains with spa type t034 do not compete for colonization space with the corresponding MRSA strains.
van Duijkeren et al. have reported that in different S. aureus ST398 pig isolates, only type IV and V (5C2) SCCmec elements could be detected (34). All strains investigated were tetracycline resistant and did not have spa type t034. In contrast, our sequencing results show that the SCCmec from the tetracycline-sensitive ST398 strain UMCG-M4 is a type V (5C2&5)-like SCCmec element that shares a similar overall structure with the SCCmec of an ST59 S. aureus isolate from Taiwan and the SCCmec of S. pseudintermedius (19). However, DNA of the ST59 strain could be digested with SmaI. This difference implies that if the ST59 strain does have a functional restriction-modification system, it will be different from the one(s) in our ST398 strains. The PFGE-untypeable phenotype and the presence of the hsd restriction-modification system in the SCCmec of strain UMCG-M4 indicates a possible involvement of this system in the DNA methylation at the CCNGG sequence (2). A DNA comparison of the known type V (5C2&5)-like SCCmec elements from S. aureus PM1, S. pseudintermedius, and S. aureus UMCG-M4 shows that these cassettes contain several insertions, deletions, and mutations (Fig. 3). These differences reveal the presence of different subtypes of the type V (5C2&5) SCCmec. Once the complete sequences of all these cassettes are available, a precise subtype classification can be performed in accordance with the new guidelines on the classification of SCCmec elements as proposed by the International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (20).
The present findings show that parts of the type V (5C2&5) cassettes are highly similar to sequences within SCCmec elements from S. haemolyticus and S. pseudintermedius. Recent studies of the structures of SCCmec elements in methicillin-resistant variants of the coagulase-negative staphylococci (CoNS) Staphylococcus epidermidis and S. haemolyticus showed the presence of ccrC-carrying units flanking the mec gene complex (28). This could indicate that the type V (5C2&5) SCCmec from strain UMCG-M4 and the SCCmec elements from CoNS share a history. Clearly, these findings underpin the view that SCCmec elements have a mosaic structure and that certain parts of these cassettes have a high tendency for intra- and interspecies recombination. The mechanisms by which such rearrangements occur are largely undefined, but the results of our present study suggest that homologous recombination can play an important role in these processes. The interspecies interactions leading to new SCCmec variants are of special clinical interest, because these hybrid cassettes are difficult to recognize in standard PCR-based diagnostic assays.
The observed in vivo MRSA-to-MSSA conversion in our ST398 strains occurred by recombination between ccrC1 allele 8 and ccrC1 allele 10. This mechanism for MRSA-to-MSSA conversion has not been reported before. Previous studies have revealed site-specific chromosomal excision of SCCmec elements from the genome or partial deletion of mec gene complexes involving IS431 elements (26). In marked contrast to our present findings, it has been reported that MRSA-to-MSSA conversion in S. aureus PM1 is due to a complete excision of the type V (5C2&5) SCCmec from the chromosome, despite the fact that two similar ccrC-carrying units are present in this cassette (19). At present, we do not know the molecular basis for these different MRSA-to-MSSA conversion mechanisms in S. aureus PM1 and our S. aureus ST398, but this may relate to specific sequence differences between the respective type V (5C2&5) SCCmec elements. In this light, the proposal by de Lencastre et al. (6) that the presence of non-mecA-containing cassettes in the S. aureus genome might represent a platform for the acquisition of “new” features that improve the fitness of this opportunistic pathogen is very intriguing. The in vivo detection of such “empty” cassettes in our ST398 isolates may thus represent a snapshot of such an exchange event, which is often overlooked due to the simple fact that the MRSA phenotype is the most relevant for antibiotic therapy (7).
Finally, the type V (5C2&5) SCCmec of strain UMCG-M4 turned out to be remarkably stable in vitro, which raises the question of why it is observed to be unstable in vivo in not just one but at least two patients. The only clue that we currently have is that temperature seems to be a factor that can trigger the observed MRSA-to-MSSA conversion, at least in vitro. Whether the body temperature of the patients, who did suffer from fever, had an influence in the MRSA-to-MSSA conversion is difficult to answer retrospectively. Clearly, there are many other factors playing a role in staphylococcal propagation and survival in and on the human body which may have played a role in the loss of the mec complex via recombination between the two ccrC genes. It will be a major challenge for future research to identify the conditions that trigger MRSA-to-MSSA conversion in vivo. An improved understanding of such conversion mechanisms may ultimately yield new and improved strategies to combat MRSA.
Supplementary Material
Acknowledgments
We thank Willy Baas and Wietske Postma for performing the PFGE and Jan Pieter van der Berg for expert technical assistance.
M.A.C., S.K., J.P.A., S.E., H.G., K.O., G.B., and J.M.V. were supported in part by CEU project grants LSHG-CT-2004-503468, LSHG-CT-2004-005257, LSHM-CT-2006-019064, LSHG-CT-2006-037469, and PITN-GA-2008-215524; Top Institute Pharma project grant T4-213; the transnational SysMO initiative through project grant BACELL SysMO; the European Science Foundation under the EUROCORES Programme EuroSCOPE; and grant 04-EScope 01-011 from the Research Council for Earth and Life Sciences of the Netherlands Organization for Scientific Research. S.E. was supported by DFG research grants SFB/TR34 and FOR585. K.O. was supported by DFG research grants SFB/TR34 and SFB630.
Footnotes
Published ahead of print on 7 December 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Aires-de-Sousa, M., K. Boye, H. de Lencastre, A. Deplano, M. C. Enright, J. Etienne, A. Friedrich, D. Harmsen, A. Holmes, X. W. Huijsdens, A. M. Kearns, A. Mellmann, H. Meugnier, J. K. Rasheed, E. Spalburg, B. Strommenger, M. J. Struelens, F. C. Tenover, J. Thomas, U. Vogel, H. Westh, J. Xu, and W. Witte. 2006. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J. Clin. Microbiol. 44:619-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bens, C. C., A. Voss, and C. H. Klaassen. 2006. Presence of a novel DNA methylation enzyme in methicillin-resistant Staphylococcus aureus isolates associated with pig farming leads to uninterpretable results in standard pulsed-field gel electrophoresis analysis. J. Clin. Microbiol. 44:1875-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chongtrakool, P., T. Ito, X. X. Ma, Y. Kondo, S. Trakulsomboon, C. Tiensasitorn, M. Jamklang, T. Chavalit, J. H. Song, and K. Hiramatsu. 2006. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob. Agents Chemother. 50:1001-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dall'Antonia, M., P. G. Coen, M. Wilks, A. Whiley, and M. Millar. 2005. Competition between methicillin-sensitive and -resistant Staphylococcus aureus in the anterior nares. J. Hosp. Infect. 61:62-67. [DOI] [PubMed] [Google Scholar]
- 5.Daurel, C., A. L. Prunier, F. Chau, L. Garry, R. Leclercq, and B. Fantin. 2007. Role of hypermutability on bacterial fitness and emergence of resistance in experimental osteomyelitis due to Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 51:344-349. [DOI] [PubMed] [Google Scholar]
- 6.de Lencastre, H., D. Oliveira, and A. Tomasz. 2007. Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power. Curr. Opin. Microbiol. 10:428-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deplano, A., P. T. Tassios, Y. Glupczynski, E. Godfroid, and M. J. Struelens. 2000. In vivo deletion of the methicillin resistance mec region from the chromosome of Staphylococcus aureus strains. J. Antimicrob. Chemother. 46:617-620. [DOI] [PubMed] [Google Scholar]
- 8.Deurenberg, R. H., and E. E. Stobberingh. 2008. The evolution of Staphylococcus aureus. Infect. Genet. Evol. 8:747-763. [DOI] [PubMed] [Google Scholar]
- 9.Donnio, P.-Y., D. C. Oliveira, N. A. Faria, N. Wilhelm, A. Le Coustumier, and H. de Lencastre. 2005. Partial excision of the chromosomal cassette containing the methicillin resistance determinant results in methicillin-susceptible Staphylococcus aureus. J. Clin. Microbiol. 43:4191-4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster, T. J. 2004. The Staphylococcus aureus “superbug.” J. Clin. Invest. 114:1693-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gally, D. L., J. A. Bogan, B. I. Eisenstein, and I. C. Blomfield. 1993. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J. Bacteriol. 175:6186-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giraud, A., I. Matic, M. Radman, M. Fons, and F. Taddei. 2002. Mutator bacteria as a risk factor in treatment of infectious diseases. Antimicrob. Agents Chemother. 46:863-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gotz, F., and B. Schumacher. 1987. Improvements of protoplast transformation in Staphylococcus carnosus. FEMS Microbiol. Lett. 40:285-288. [Google Scholar]
- 16.Guardabassi, L., M. Steggerb, and R. Skovb. 2007. Retrospective detection of methicillin resistant and susceptible Staphylococcus aureus ST398 in Danish slaughter pigs. Vet. Microbiol. 122:384-386. [DOI] [PubMed] [Google Scholar]
- 17.Hanssen, A. M., and J. U. Ericson Sollid. 2006. SCCmec in staphylococci: genes on the move. FEMS Immunol. Med. Microbiol. 46:8-20. [DOI] [PubMed] [Google Scholar]
- 18.Heusser, R., M. Ender, B. Berger-Bachi, and N. McCallum. 2007. Mosaic staphylococcal cassette chromosome mec containing two recombinase loci and a new mec complex, B2. Antimicrob. Agents Chemother. 51:390-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higuchi, W., T. Takano, L. J. Teng, and T. Yamamoto. 2008. Structure and specific detection of staphylococcal cassette chromosome mec type VII. Biochem. Biophys. Res. Commun. 377:752-756. [DOI] [PubMed] [Google Scholar]
- 20.International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53:4961-4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katayama, Y., T. Ito, and K. Hiramatsu. 2001. Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of IS431-mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin-resistant Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 45:1955-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozitskaya, S., S. H. Cho, K. Dietrich, R. Marre, K. Naber, and W. Ziebuhr. 2004. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect. Immun. 72:1210-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMahon, M. A., J. Xu, J. E. Moore, I. S. Blair, and D. A. McDowell. 2007. Environmental stress and antibiotic resistance in food-related pathogens. Appl. Environ. Microbiol. 73:211-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noto, M. J., P. M. Fox, and G. L. Archer. 2008. Spontaneous deletion of the methicillin resistance determinant, mecA, partially compensates for the fitness cost associated with high-level vancomycin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 52:1221-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 28.Ruppe, E., F. Barbier, Y. Mesli, A. Maiga, R. Cojocaru, M. Benkhalfat, S. Benchouk, H. Hassaine, I. Maiga, A. Diallo, A. K. Koumare, K. Ouattara, S. Soumare, J. B. Dufourcq, C. Nareth, J. L. Sarthou, A. Andremont, and R. Ruimy. 2009. Diversity of staphylococcal cassette chromosome mec structures in methicillin-resistant Staphylococcus epidermidis and Staphylococcus haemolyticus strains among outpatients from four countries. Antimicrob. Agents Chemother. 53:442-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaaff, F., A. Reipert, and G. Bierbaum. 2002. An elevated mutation frequency favors development of vancomycin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 46:3540-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stam-Bolink, E. M., D. Mithoe, W. H. Baas, J. P. Arends, and A. V. Moller. 2007. Spread of a methicillin-resistant Staphylococcus aureus ST80 strain in the community of the northern Netherlands. Eur. J. Clin. Microbiol. Infect. Dis. 26:723-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takano, T., W. Higuchi, T. Otsuka, T. Baranovich, S. Enany, K. Saito, H. Isobe, S. Dohmae, K. Ozaki, M. Takano, Y. Iwao, M. Shibuya, T. Okubo, S. Yabe, D. Shi, I. Reva, L. J. Teng, and T. Yamamoto. 2008. Novel characteristics of community-acquired methicillin-resistant Staphylococcus aureus strains belonging to multilocus sequence type 59 in Taiwan. Antimicrob. Agents Chemother. 52:837-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeuchi, F., S. Watanabe, T. Baba, H. Yuzawa, T. Ito, Y. Morimoto, M. Kuroda, L. Cui, M. Takahashi, A. Ankai, S. Baba, S. Fukui, J. C. Lee, and K. Hiramatsu. 2005. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 187:7292-7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Belkum, A., D. C. Melles, J. K. Peeters, W. B. van Leeuwen, D. E. van Duijkeren, X. W. Huijsdens, E. Spalburg, A. J. de Neeling, H. A. Verbrugh, and Dutch Working Party on Surveillance and Research of MRSA-SOM. 2008. Methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398 in pigs and humans. Emerg. Infect. Dis. 14:479-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Duijkeren, E., R. Ikawaty, M. J. Broekhuizen-Stins, M. D. Jansen, E. C. Spalburg, A. J. de Neeling, J. G. Allaart, A. van Nes, J. A. Wagenaar, and A. C. Fluit. 2008. Transmission of methicillin-resistant Staphylococcus aureus strains between different kinds of pig farms. Vet. Microbiol. 126:383-389. [DOI] [PubMed] [Google Scholar]
- 35.van Loo, I. H., B. M. Diederen, P. H. Savelkoul, J. H. Woudenberg, R. Roosendaal, A. van Belkum, N. Lemmens-den Toom, C. Verhulst, P. H. van Keulen, and J. A. Kluytmans. 2007. Methicillin-resistant Staphylococcus aureus in meat products, The Netherlands. Emerg. Infect. Dis. 13:1753-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Loo, I., X. Huijsdens, E. Tiemersma, A. de Neeling, N. van de Sande-Bruinsma, D. Beaujean, A. Voss, and J. Kluytmans. 2007. Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg. Infect. Dis. 13:1834-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voss, A., F. Loeffen, J. Bakker, C. Klaassen, and M. Wulf. 2005. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 11:1965-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welinder-Olsson, C., K. Floren-Johansson, L. Larsson, S. Oberg, L. Karlsson, and C. Ahren. 2008. Infection with Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus t034. Emerg. Infect. Dis. 14:1271-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Witte, W., B. Strommenger, C. Stanek, and C. Cuny. 2007. Methicillin-resistant Staphylococcus aureus ST398 in humans and animals, Central Europe. Emerg. Infect. Dis. 13:255-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wulf, M., A. van Nes, A. Eikelenboom-Boskamp, J. de Vries, W. Melchers, C. Klaassen, and A. Voss. 2006. Methicillin-resistant Staphylococcus aureus in veterinary doctors and students, The Netherlands. Emerg. Infect. Dis. 12:1939-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wulf, M. W., E. Tiemersma, J. Kluytmans, D. Bogaers, A. C. Leenders, M. W. Jansen, J. Berkhout, E. Ruijters, D. Haverkate, M. Isken, and A. Voss. 2008. MRSA carriage in healthcare personnel in contact with farm animals. J. Hosp. Infect. 70:186-190. [DOI] [PubMed] [Google Scholar]
- 42.Yu, F., Z. Chen, C. Liu, X. Zhang, X. Lin, S. Chi, T. Zhou, Z. Chen, and X. Chen. 2008. Prevalence of Staphylococcus aureus carrying Panton-Valentine leukocidin genes among isolates from hospitalised patients in China. Clin. Microbiol. Infect. 14:381-384. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.