Abstract
Commensal bacteria possess immunostimulatory activities that can modulate host responses to affect development and homeostasis in the intestine. However, how different populations of resident bacteria stimulate the immune system remains largely unknown. We characterized here the ability of intestinal and oral microflora to stimulate individual pattern recognition receptors (PRRs) in bone marrow-derived macrophages and mesothelial cells. The intestinal but not oral microflora elicited age- and cell type-specific immunostimulation. The immunostimulatory activity of the intestinal microflora varied among individual mice but was largely mediated via Toll-like receptor 4 (TLR4) during breast-feeding, whereas it became TLR4 independent after weaning. This transition was associated with a change from a microflora rich in TLR4-stimulatory proteobacteria to one dominated by Bacteroidales and/or Clostridiales that poorly stimulate TLR4. The major stimulatory activity of the intestinal microflora was still intact in NOD1-, NOD2-, TLR2-, TLR4-, TLR5-, TLR9-, TLR11-, ASC-, or RICK-deficient cells but still relied on the adaptor MyD88. These studies demonstrate a transition in the intestinal microflora accompanied by a dynamic change of its ability to stimulate different PRRs which control intestinal homeostasis.
Accumulating evidence indicates that environmental bacteria can regulate the development and homeostasis of the host immune system, particularly within the gut, and affect susceptibility to a variety of diseases (3, 5, 6, 9, 10, 40, 44). Both humans and animals harbor a large number of nonpathogenic residential bacteria, especially in the intestine and oral cavity (41). Uncontrolled translocation of bacteria or bacterial components into systemic tissues of the host often results in bacteremia and sepsis (8), which causes significant mortality worldwide each year. On the other hand, intestinal bacteria contain immunostimulatory molecules that can regulate local immunity, epithelial development, immunotolerance, and susceptibility to inflammatory bowel disease (2, 41). The bacterium-derived molecules are recognized by innate immune receptors, including Toll-like receptors (TLRs) and Nod-like receptors (NLRs) (8, 20). TLRs and NLRs, often referred as pattern recognition receptors (PRRs), are involved in the recognition of commensal and pathogenic bacteria, as well as in the clearance of pathogens through interaction with their cognate microbial molecules (8, 20). Interactions between PRRs and commensal bacteria have been demonstrated to be important for gut homeostasis. MyD88, an essential adaptor for TLR signaling, has been shown to be important for epithelial homeostasis (40) and IgA secretion in the intestine (3, 44), and TLR9 has been shown to be important for the balance of regulatory T/Th17/Th1 cells (10). In addition, NOD1, an NLR family member, was shown to play a role in the development of intestinal lymphoid tissue via the recognition of commensal bacteria (5). Finally, genetic variation in NOD1 affects the susceptibility to allergic disease (17, 49) and Crohn's disease (31) and that in NOD2 regulates susceptibility to Crohn's disease (11, 16, 36) and graft-versus-host disease (14). In the oral cavity, the immunostimulatory properties of periodontal bacteria are believed to be important for the development of dental diseases (47).
Although beneficial interactions between commensal bacteria and innate immune receptors have been demonstrated, the dominant bacterial species that are responsible for PRR stimulation in the normal intestine and oral cavity are unknown. Moreover, the commensal-dependent immunostimulatory activity for the various innate immune receptors has not been characterized. In humans, both genetic factors and the environment, including the diet, modulate the composition of the microflora (7, 15, 23, 33, 41, 48). Consequently, the intestinal microflora of individual humans is highly diverse (7, 15, 23, 33, 41). By taking advantage of the mouse model which allows a more uniform environmental and genetic platform, we analyzed the immunostimulatory activity induced by the oral and intestinal microflora and linked the activity to specific populations of commensal bacteria that emerge during postnatal development. We study provide here a comprehensive resource on microflora analysis in the mouse that is expected to facilitate future studies of the interaction between commensal bacteria and host immunity.
MATERIALS AND METHODS
Reagents and bacteria.
MDP, synthetic bacterial lipoprotein (sBLP; Pam3-Cys-OH), and Escherichia coli O55:B5 lipopolysaccharide (LPS) were purchased from Sigma-Aldrich (St. Louis, MO). The NOD1-stimulatory compounds, KF1B and iE-DAP, were described previously (30). LPS was confirmed to be free of contamination with NOD1 and NOD2 stimulatory activity, as detected by an HEK293T bioassay (described previously [12]). E. coli K-12, Bacteroides fragilis NCTC10581, B. vulgatus ATCC 8482, Lactobacillus plantarum ATCC 8041, and L. pentosus ATCC 11580 were cultured as described previously (12). The expression plasmids of NOD1, NOD2, TLR4, and MD2 were described previously (12).
Mice.
Wild-type (WT) C57BL/6 (B6) and BALB/c mice, as well as BALB/c mice deficient in TLR4, were obtained from the Jackson Laboratory (Bar Harbor, ME). B6 mice deficient in ASC, MyD88/TRIF, NOD1, RICK, TLR2, and TLR4 were described in previous studies (12, 30, 45, 52). MyD88/RICK-deficient mice in B6 background were provided by G. Cheng (University of California at Los Angeles). TLR9-deficient BALB/c mice and an 8-week-old germfree B6 male mouse were kindly provided by T. J. Standiford and K. Eaton (University of Michigan, Ann Arbor), respectively. Bone marrow-derived macrophages (BMM) from TLR5- and TLR11-deficient B6 mice were kindly provided by Andrew Gewirtz (Emory University, Atlanta). All mice were housed, bred, and fed Laboratory Rodent Diet 5001 (Labdiet, Purina Mills) under specific-pathogen-free conditions in the animal facility of the University of Michigan. Mice were given food and water ad libitum in an environmentally controlled room with a cycle of 12 h of light and 12 h of darkness. The mouse studies were approved by the University of Michigan Committee on Use and Care of Animals.
Preparation of homogenates from oral and intestinal organs, bacteria counting.
Microflora was analyzed in B6 mice housed in different cages (seven to 10 mice per time point) at the same room of the animal facility. All mice used for microflora analysis were the first generation from parents obtained from Jackson Laboratory and raised from one pair of parents per cage. All oral and intestinal organs were isolated from 3-, 7-, and 21-day-old wild-type B6 mice. Whole small intestines, ceca, and colons were isolated and weighed. After the removal of skin, the apical part of the jaw joint between the mandible and the maxilla was isolated and weighed. The organs were homogenized in 1 ml of phosphate-buffered saline, frozen, and stored at −80°C until analyses.
LAL assay and NOD1-, NOD2-, and TLR4-stimulatory HEK293T bioassay.
The quantity of LPS in homogenized tissue samples was determined by using Limulus amebocyte lysate (LAL) assay kit (Lonza, Walkersville, MD) in accordance with the manufacturer's instructions. E. coli O55:B5 LPS was used as a standard. The NOD1-, NOD2-, and TLR4-stimulatory activities were determined by HEK293T bioassay as described previously (12). Briefly, HEK293T cells were transfected with the NOD1, NOD2, or TLR4+MD2 expression plasmids by the calcium phosphate method and, at 8 h posttransfection, the cells were treated with medium containing series of specific ligands (iE-DAP for NOD1, MDP for NOD2, and E. coli O55:B5 LPS for TLR4) or tested samples. At 24 h posttransfection, ligand-dependent NF-κB activation was determined with a reporter assay. The bioactivity was calculated from a comparison between the samples, and the titer curve of the NF-κB activation levels was obtained by using known amounts of specific ligands. One stimulatory unit was defined as the level of immunostimulation obtained with 1 ng of each ligand, as described previously (12).
16S rRNA gene analysis, DGGE, and quantitative analysis of bacteria.
The 16S rRNA gene (16S rRNA gene) was amplified by 35 cycles of PCR at 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min, using the broad-range bacterial primers CCAAACTCCTACGGGAGGCAGCAG and CATGGACTACCAGGGTATCTAATC (12) from the bacterial DNA pool (1/50 total DNA) from each mouse organ. This first PCR step was essential to avoid the contamination of 18S mouse rRNA gene in bacterial 16S rRNA gene pools amplified from samples, which contain very low numbers of bacteria. Amplified products were verified by agarose gel electrophoresis and purified by QIAEX II (Qiagen). The PCR products were then ligated into pGEM-T (Promega) for sequencing analysis or used as templates for denaturing gradient gel electrophoresis (DGGE) analysis. For the results shown in Table S1 in the supplemental material, the pGEM-T 16S rRNA gene library was introduced into E. coli STBL2 (Invitrogen). The cloned 16S rRNA gene fragments amplified by 25 cycles of PCR (94°C for 1 min, 60°C for 1 min, and 72°C for 1 min) with T7 and SP6 primers were purified from agarose gel by QIAEX II after electrophoresis and were sequenced from both ends using the same primers. The resulted sequences were subjected to an online BLASTN analysis (National Center for Biotechnology Information, National Institutes of Health) for bacterial identification. For DGGE analysis, we conducted nested PCR as follows. In the first PCR, samples were initially PCR amplified by using the above-mentioned broad-range bacterial primers in a reaction volume of 25 μl. PCR amplification was carried out using the following program: 2 min at 94°C; 30 cycles of 1 min at 94°C, 1 min at 56°C, and 1 min at 72°C; and 5 min at 72°C. The first PCR products were diluted 1/1,000 with Tris-EDTA (TE) buffer and used as a template for a second PCR with 518r and GC-clumped 341f primers as described previously (34). The second PCR was performed 10 cycles of touchdown PCR (denaturation at 94°C for 1 min, annealing for 1 min with 1°C/cycle decrement from 65 to 56°C, and elongation at 72°C for 1 min), followed by 15 cycles of regular PCR (1 min at 94°C, 1 min at 55°C, and 1 min at 72°C) after 94°C for 5 min, and a final elongation for 30 min at 72°C. The PCR products were purified with a commercial kit (Promega).
DGGE analysis was performed with a D-code system (Bio-Rad) as described previously (34). Individual DNA fragments extracted from the gel were amplified by PCR with the primers 341f and 518r, subcloned into the pGEM-T vector, and sequenced as described above. The homogeneity of the DNA fragments was confirmed by sequencing multiple clones.
To determine the total bacteria, the bacteria (Gram positive and Gram negative) in homogenates were counted after Gram staining. To determine the DNA amounts of enterobacteria, quantitative PCR was performed by using universal 16S rRNA gene primers Eco1457F/Eco1652R(4) with various cycles at 94°C for 1 min, 65°C for 1 min, and 72°C for 1 min on an ABI Prism 7900HT system (Applied Biosystems, Foster City, CA). The DNA amounts were determined from standard curves established by quantitative PCR on serial dilutions of reference E. coli DNA and normalized by total bacterial DNA amount estimated by using 341f and 518r (34).
Immunostimulation assay for macrophages and mesothelial cells.
BMM and mesothelial cells were prepared and cultured as described previously (12). The homogenized tissue samples were heated at 98°C for 30 min. Samples diluted 100-fold were incubated with 1 × 105 BMM or 2 × 104 mesothelial cells in 0.2 ml of medium. After 12 h of stimulation, levels of IL-6, CCL5, CXCL1, and interleukin-10 (IL-10) in the medium were determined by sandwich ELISA kits (BD Pharmingen, San Diego, CA).
Statistical analysis.
The coefficient of determination (R2) was used to assess the correlation between levels of cytokine secretion, bacterial numbers, and LPS content in mouse samples. Differences in cytokine/chemokine production between wild-type (WT) and mutant cells were determined by using a two-tailed t test with unequal variance (Aspin-Welch's t test; Excel, Microsoft).
RESULTS
Accumulation of immunostimulatory activity in the oral and intestinal microflora during mouse development.
To determine the immunostimulatory activity of the oral and intestinal bacterial populations, BMM were stimulated with heat-inactivated tissue homogenates prepared from the oral cavity and intestinal sites. Intestinal homogenates was used in order to include in our analysis both nonadherent luminal and tissue-adherent bacteria. As a control, BMM were also stimulated with intestinal homogenates from a germfree adult mouse. As expected, IL-6 production was absent when BMM were stimulated with germfree intestinal homogenates (Fig. 1A). Stimulation of BMM with cecal and colonic homogenates from 21-day-old mice, however, resulted in the secretion of IL-6, CCL5 (RANTES), and IL-10 (Fig. 1A, B, and C). In contrast, no significant immunostimulatory activity was observed with oral or small intestinal homogenates from mice at any age (Fig. 1A, B, and C), with the exception of a single mouse which possessed a high percentage of E. coli within the small intestine (mouse 7W02, see also Fig. 3B, 4C, and 5B). The overall profiles for IL-6, CCL5 and IL-10 production were similar (R2 = 0.93 for IL-6 and CCL5; R2 = 0.88 for IL-6 and IL-10; R2 = 0.80 for CCL5 and IL-10) (Fig. 1D), suggesting no preferential induction of proinflammatory or anti-inflammatory responses. The secretion levels of these cytokines were also weakly associated with bacterial number (R2 = 0.51 for CCL5; R2 = 0.55 for IL-10) (Fig. 1E), suggesting that a factor other than total bacterial number is responsible for the observed differences in the cytokine production.
FIG. 1.
Stimulation of BMM by oral and intestinal microflora. BMM were stimulated with tissue homogenates derived from the oral cavity, small intestine (SI), and colon from different 3-, 7-, and 21-day-old mice (n = 7) or adult germfree (gf) mice. The amounts of IL-6 (A), CCL5 (B), and IL-10 (C) in culture supernatants were determined by enzyme-linked immunosorbent assay (ELISA). (D) Correlation between IL-6 and CCL5 (•) levels and between IL-6 and IL-10 levels (○). (E) Correlation between total bacterial number and levels of CCL5 (•) and IL-10 (○). (F) Secretion of CXCL1 by mesothelial cells stimulated with tissue homogenates from the indicated sites from 3-, 7-, and 21-day-old B6 mice. The amounts of CXCL1 in culture supernatants were determined by ELISA. Each symbol represents mean value of triplicate cultures from individual mice. The standard deviations (SD) of triplicate cultures for individual mice averaged less than 19%. Bars denote mean values. *, P < 0.001.
FIG. 3.
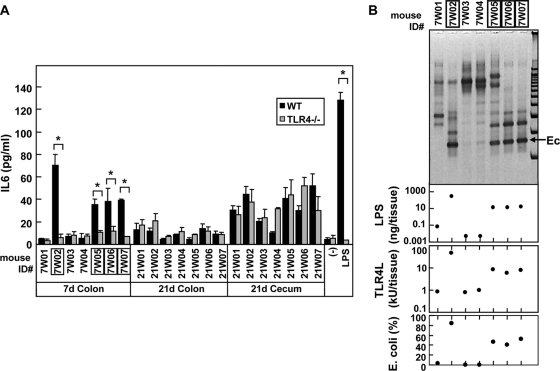
TLR4-dependent macrophage-stimulatory activity in 7-day-old mice is associated with E. coli predominance. (A) BMM from WT and TLR4−/− mice were treated with the medium alone (−) and heat-inactivated homogenates (diluted 100-fold) from the indicated tissues of 3-, 7-, and 21-day-old B6 mice. Stimulation with 100 ng of LPS/ml is also shown as a control. The amounts of IL-6 secreted by BMM after 12 h of stimulation were determined by ELISA. The data are represented as means ± the SD. *, P < 0.001. (B) Comparison of dominant bacteria LPS content and TLR4-stimulatory activity in the small intestinal microflora on day 7. The major bacterial species from individual mice were analyzed by DGGE (upper panel) and compared to the LPS content and TLR4-stimulatory activity (second and third panels). Ec, band corresponding to E. coli as determined by sequencing. The percentage of E. coli in the whole bacterial population of the colon(bottom panel) was determined by real-time PCR as described in Materials and Methods. The numbers of mice that possessed E. coli dominant microflora are boxed.
FIG. 4.
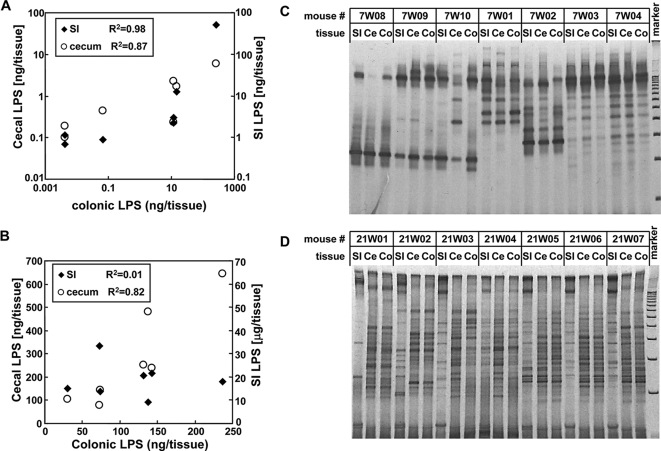
Transition of dominant bacteria in intestinal microflora during mouse development. (A and B) Correlation between LPS levels within the small intestine (SI), cecum (Ce), and colon (Co) on day 7 (A) and day 21 (B). (C and D) Comparison of dominant bacterial species in the intestine by DGGE analysis on day 7 (C) and day 21 (D). All mouse sets used in panels A and B were the same as those used in panels C and D, respectively.
FIG. 5.
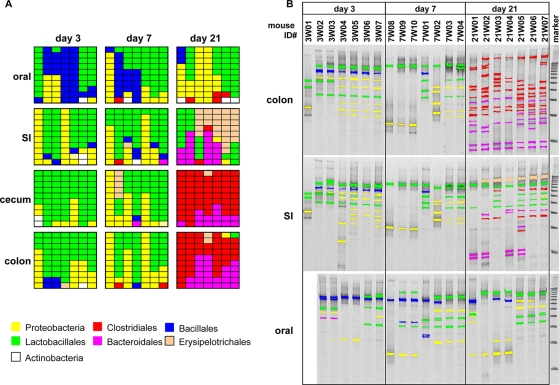
Identification of dominant bacteria in oral and intestinal microflora during mouse development. (A) The bacterial DNA prepared from homogenates of oral and intestinal tissues were amplified with 16S rRNA gene consensus primer set. A total of 876 clones were randomly isolated and sequenced from 16S rRNA gene libraries generated from each microflora. The identity and homology of each clone were determined by using the BLASTN program. First, 10 clones from each sample are shown by the distinct colors corresponding to the taxonomic groups. The full description of bacteria species are shown in Table S1 in the supplemental material. (B) The DNA was further amplified with the GC-clump-linked consensus primer and separated by DGGE with DNA 100-bp ladder markers. The major DNA fragments were recovered from the gel and sequenced as described in Materials and Methods. The identities of bacteria were determined by homology search using the BLASTN program. The DNA fragments of characterized bacteria are depicted in the same colors as in panel A. The original images and full descriptions of the bacterial species are shown in Fig. S2 and Table S2, respectively, in the supplemental material.
Nonhematopoietic cells are also important for host defense through the production of chemokines and other antimicrobial molecules. Since mesothelial cells, but not macrophages, can respond to certain microbial molecules such as NOD1 agonists (30), we used primary mesothelial cells to assess the response induced by intestinal homogenates. Whereas there was no detectable BMM-immunostimulatory activity associated with small intestinal homogenates on day 21 (Fig. 1A to C), stimulation of mesothelial cells by either small intestinal or colonic homogenates resulted in significant secretion of CXCL1 (Fig. 1F). These results suggest that small intestinal microflora from 21-day-old mice possesses cell-type-specific stimulatory activity.
Age-specific TLR4-stimulatory activity of LPS in the oral and intestinal microflora.
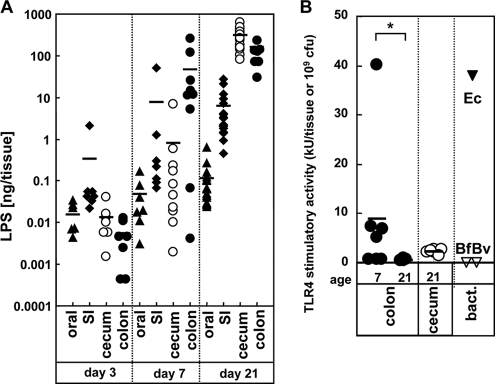
A major bacterial component capable of inducing inflammatory and other immune responses is LPS. We used the LAL assay to determine the amount of LPS in the mouse oral cavity and intestine. The LPS content in the oral tissue was very low (mean, <1 ng/tissue) compared to that found in the intestinal tract (Fig. 2A). From day 3 to day 21 after birth, the amount of LPS in the oral and intestinal tissue increased ∼300-fold (Fig. 2A), a finding consistent with the observed rise in the number and density of bacteria (see Fig. S1 in the supplemental material). In line with these results, the small intestine contained the majority of LPS early during development (Fig. 2A). Notably, the amount of LPS in the colon and cecum was only 1.5 and 1.6% of the total LPS on day 3, but this increased to 35 and 65%, respectively, by day 21 (Fig. 2A). Since LPS is only present in Gram-negative bacteria, these results suggest that the small intestine is the major site of Gram-negative bacteria early on during development (day 3), but by day 21, these organisms became more common in the cecum and colon.
FIG. 2.
Difference between the LPS content and TLR4-immunostimulatory activity in oral and intestinal tissues. (A) Total amounts of LPS on day 3, 7, and 21 in oral tissue (▴), small intestine (SI; ⧫), cecum (○), and colon (•) were determined by LAL assay. (B) TLR4-stimulatory activity in oral and intestinal tissues. The levels of TLR4-stimulatory activity in cecal and colonic homogenates (n = 7) at days 7 and 21 were determined by the bioassay using HEK293T cells expressing TLR4 and MD-2. Bars denote mean values. As reference controls, the LPS levels of 109 CFU of Gram-negative bacteria, E. coli (Ec), B. fragilis (Bf), and B. vulgatus (Bv) are shown. *, P < 0.001.
The LAL assay detects the amount of lipid X moiety in LPS but not TLR4-specific stimulatory activity (46). To determine whether the ability of intestinal LPS to stimulate TLR4, we used a TLR4 reporter assay in HEK293T cells (12). Surprisingly, colon homogenates harvested on day 21, which were rich in total LPS, showed little TLR4/MD2-stimulatory activity (Fig. 2B [see details below] and Fig. 3). As a reference, E. coli, but not two Gram-negative Bacteroidetes species, possessed strong TLR4-stimulatory activity (Fig. 2B). TLR4-induced stimulation by cecal homogenates was also very low on day 21. However, on day 7, there was significant TLR4-stimulatory activity, although there was wide variability among the individual mice tested at this age (Fig. 2B). These results indicate that on day 21 after birth, the LPS derived from Gram-negative bacteria in the cecum and colon is poorly TLR4 stimulatory.
Transition of immunostimulatory activity from TLR4 dependent to TLR4 independent during the intestinal microflora development.
Because the colonic microflora from a subgroup of 7-day-old mice possessed a significant level of TLR4-stimulatory activity (Fig. 2B), we next determined whether the cytokine production by BMM in response to colonic microflora was mediated by TLR4. Using BMM from TLR4-deficient and control WT mice, we found that colonic homogenates from 7-day-old mice possessing significant TLR4-stimulatory activity (mice7W02, 7W05, 7W06, and 7W07) failed to induce any significant cytokine production in TLR4-deficient BMM (Fig. 3A). Furthermore, the immunostimulatory activity of the colonic microflora in individual mice on day 7 correlated with both LPS content and TLR4-stimulatory levels identified by using the TLR4 reporter bioassay (second and third panels, Fig. 3B). Denaturing gradient gel electrophoresis (DGGE)-coupled sequencing analysis also revealed that E. coli contributed to the majority of LPS in the colon on day 7 (top panel in Fig. 3B). Furthermore, quantitative PCR analysis using specific primers for enterobacteria showed a direct correlation between the relative percentage of E. coli and TLR4-stimulatory activity on day 7 (bottom panel of Fig. 3B) and on day 21 when the levels of E. coli in the cecum and colon are very low (0.01% ± 0.01% and 0.03% ± 0.05%, respectively). Collectively, these results indicate that E. coli is a major source of TLR4-dependent immunostimulatory activity in the colonic microflora and is responsible for the TLR4-dependent cytokine responses observed in the intestine of 7-day-old mice. In contrast, BMM responses to colonic and cecal homogenates from 21-day-old mice were independent of TLR4 (Fig. 3A). Thus, the colonic microflora after the weaning stage consists of bacteria that stimulate BMM in a TLR4-independent manner.
Developmental transition of the intestinal microflora.
The above experiments suggest that bacterial numbers, LPS content and TLR4-stimulatory activity in the intestinal tract are site and age specific. When we compared the LPS content in the small intestines, ceca, and colons of individual mice, we found a direct correlation between the three tissues on day 7 (R2 = 0.98, colon versus small intestine; R2 = 0.87 colon versus cecum, Fig. 4A). However, although the LPS content in the cecum directly correlated with that found in the colon on day 21 (R2 = 0.82), there was no correlation between the colon and small intestine (R2 = 0.01, Fig. 4B). These results suggest that, on day 7, the microflora is similar at different sites of the intestine but diverges between the small intestine and cecum/colon by day 21.
To identify the basis for the change in LPS content between the small and large intestine during mouse development, we investigated the dominant bacterial populations within the small and large intestine by DGGE analysis. Although the electrophoretic patterns can differ between mice indicative of variable microbial communities, the bacterial composition of individual mice was similar at different intestinal sites on day 7 after birth (Fig. 4C). In contrast, on day 21, the bacterial populations within the cecum and colon were nearly identical but markedly different from that found in the small intestine (Fig. 4D). Thus, the dominant bacterial populations within the small intestine, cecum, and colon are relatively uniform at the breast-feeding age but diverge in the small intestine and cecum/colon after weaning.
Identification of dominant bacterial species in the oral and intestinal microflora.
To identify the bacterial sources of the observed immunostimulatory activities within the intestine, we further characterized the microflora at these tissue sites by 16S rRNA gene-based taxonomy. First, we characterized 876 clones randomly isolated from a bacterial DNA library generated by PCR using 16S rRNA gene consensus primers to estimate relative proportions of different bacterial species (Fig. 5A). Second, dominant bacterial species in the oral and intestinal microflora were identified by sequencing DNA fragments separated by DGGE (Fig. 5B). Based on sequencing analysis, the dominant bacterial populations in the intestinal microflora on day 3 consist of the Gram-negative Klebsiella pneumoniae-K. oxytoca-Enterobacter hormaechei subgroup of Enterobacteriaceae and Gram-positive Lactobacillales (Fig. 5 and see Fig. S2 in the supplemental material). Because these bacteria are lactose-fermenting facultative anaerobic organisms that grow well under aerobic conditions, this result suggests that these bacteria are the initial colonizers of the mouse intestine during breast-feeding.
On day 7, the dominant bacterial populations within the intestine were similar to that observed on day 3, except that some mice acquired a predominance of E. coli (see Fig. S2 and Table S2 in the supplemental material). Notably, there was a dramatic change in dominant bacterial species within the colon on day 21, which was associated with a high level of bacterial diversity (Fig. 5D). In contrast, on days 3 and 7 the intestinal microflora consisted primarily of facultative anaerobic bacteria, but on day 21 there was a switch to obligate anaerobic bacteria, namely, Clostridiales and Bacteroidales (top panel in Fig. 5B). Therefore, the transition from E. coli-rich microflora to Clostridiales/Bacteroidales-rich microflora can explain the loss of TLR4-stimulatory activity of the intestinal microflora after weaning.
The major innate immune responses induced by colonic microflora after weaning are mediated via MyD88.
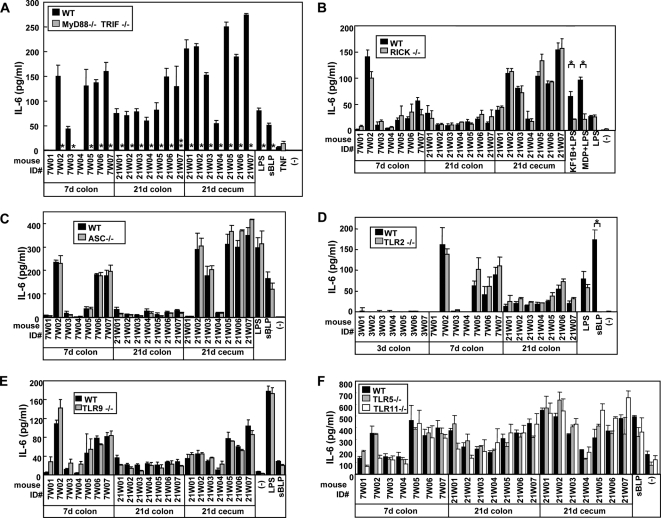
To determine whether the TLR4-independent stimulation of BMM by the colonic microflora from 21-day-old mice was dependent on TLR and/or NLR signaling, we analyzed cytokine responses in BMM from mice lacking MyD88/TRIF, RICK, and ASC, which are essential downstream mediators of TLRs, NOD1/NOD2, and NLRP3/NLRC4 signaling, respectively (Fig. 6). No difference in cytokine production was observed between WT and RICK- or ASC-deficient BMM (Fig. 6B and C). Importantly, there was no detectable IL-6 production by MyD88/TRIF-deficient BMM (Fig. 6A), indicating that the immunostimulatory activity of colonic and cecal microflora in 21-day-old mice is largely dependent on TLR signaling. However, IL-6 production was unaffected by the absence of either TLR2, TLR5, TLR9, or TLR11 (Fig. 6D to F), suggesting that there is TLR redundancy or that a TLR other than these TLRs is responsible for mediating the immunostimulatory activity of the intestinal microflora from 21-day-old mice.
FIG. 6.
Dominant role of MyD88/TRIF signaling in immune responses of BMM to intestinal microflora. BMM from a control WT, MyD88−/− TRIF−/− (A), RICK−/− (B), ASC−/− (C), TLR2−/− (D), TLR9−/− (E), TLR5−/−, or TLR11−/− (F) mice were stimulated with medium alone (−) or heat-inactivated homogenates from the indicated tissues of 7- and 21-day-old B6 mice. Stimulation with 10 ng of LPS/ml, 5 μg of KF1B/ml plus 10 ng of LPS/ml, 5 μg of MDP/ml plus 10 ng of LPS/ml, or 1 μg of sBLP/ml is shown as a control. The amounts of IL-6 in culture supernatants after 12 h of stimulation were determined by ELISA. The data are represented as means ± the SD. * and **, P < 0.001 and P < 0.01, respectively.
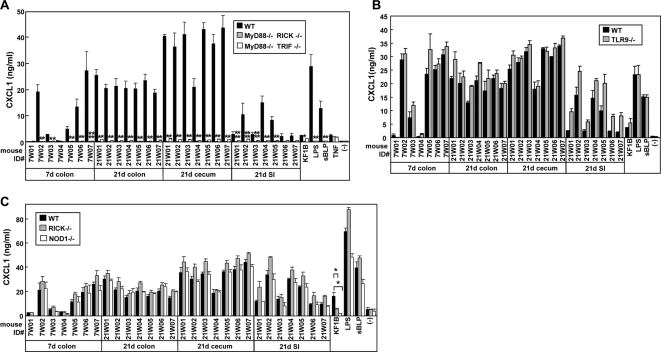
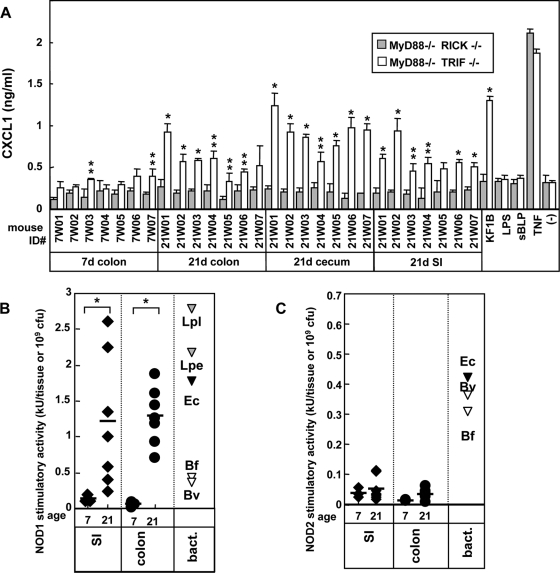
We next used mesothelial cells to assess immune response to the intestinal microflora from day 21 intestine (Fig. 7). Secretion of CXCL1 by RICK-, NOD1-, and TLR9-deficient mesothelial cells was similar to that observed in WT cells (Fig. 7B and C). Consistent with the BMM results, the innate immune response was dramatically impaired in mesothelial cells deficient in MyD88/TRIF and MyD88/RICK (Fig. 7A). This indicates that the major immunostimulatory activity found in the colon and cecum on day 21 is primarily dependent on MyD88, and not on NOD1, TLR2, or TLR9. The MyD88-deficient mesothelial cells were also impaired in their response to 21-day small intestinal homogenates, suggesting that cell-type-specific immunostimulatory activity of the small intestinal microflora is also dependent on MyD88. Interestingly, there was some residual CXCL1 secretion in MyD88/TRIF-deficient mesothelial cells, which was abrogated in MyD88/RICK-deficient mesothelial cells (Fig. 8A). The HEK293T bioassay revealed that NOD1-stimulatory activity was increased 8.9- and 23-fold in the small intestine and colon on day 21, whereas that mediated by NOD2 was very low (Fig. 8B and C). These results indicate that NOD1 contributes via RICK to the stimulatory activity induced by adult small intestine and colonic microflora.
FIG. 7.
Dominant role of MyD88 in immune responses of mesothelial cells to intestinal microflora. Mesothelial cells from WT, MyD88−/− TRIF−/−, MyD88−/− RICK−/− (A), TLR9−/− (B), and RICK−/− and NOD1−/− (C) mice were treated with medium alone (−) or heat-inactivated homogenates (diluted 100-fold) from the indicated tissues of 7- and 21-day-old B6 mice. Stimulation with 100 ng of LPS/ml, 1 μg of sBLP/ml, or 10 ng of TNF-α/ml is shown as a control. The amounts of CXCL1 in culture supernatants after 12 h of stimulation were determined by ELISA. The data are represented as mean ± the SD. * and **, P < 0.001 and P < 0.01, respectively, compared to WT and mutant cells.
FIG. 8.
Secondary contribution of RICK and NOD1 in immune responses of mesothelial cells to intestinal microflora. (A) Mesothelial cells from MyD88−/−TRIF−/− and MyD88−/− RICK−/− mice were treated with medium alone (−) or heat-inactivated homogenates (diluted 100-fold) from indicated tissue of 7- and 21-day-old B6 mice. Stimulation with 100 ng of LPS/ml, 1 μg of sBLP/ml, 5 μg of KF1B/ml, 10 ng of tumor necrosis factor alpha (TNF-α)/ml is shown as a control. The amounts of CXCL1 in culture supernatants after 12 h of stimulation were determined by ELISA. The results were derived from the same experiment shown in Fig. 7B but on a different scale to reveal differences in CXCL1 secretion between MyD88−/− TRIF−/− and MyD88−/− RICK−/− mesothelial cells. The data are represented as means ± the SD. (B and C) Levels of NOD1 and NOD2 stimulatory activities in oral and intestinal tissues. The levels of NOD1 (B) and NOD2 (C) stimulatory activity in cecal and colonic homogenates (n = 7) at days 7 and 21 were determined by a bioassay using HEK293T cells expressing NOD1 or NOD2. The NOD1 stimulatory levels of 109 CFU of the mesoDAP-type peptidoglycan-containing bacteria Lactobacillus plantarum (Lpl), L. pentosus (Lpe), E. coli (Ec), B. fragilis (Bf), and B. vulgatus (Bv) or the NOD2 stimulatory levels of 109 CFU of E. coli (Ec), B. fragilis (Bf), and B. vulgatus (Bv) are shown as controls.
DISCUSSION
The mouse model is widely used for the investigation of interactions between the host and bacteria. However, no information has been published describing the immunostimulatory activity of the mouse oral and intestinal microflora, and specifically with respect to their ability to stimulate innate immune receptors. There is also a paucity of information on the population dynamics of the bacterial microflora during mouse development. Using 16S rRNA gene sequence taxonomy and sequencing coupled with DGGE analysis, we found enormous bacterial diversity in the mouse microflora similar to what has been observed in humans. Specifically, we identified ∼200 dominant bacterial species and/or phylotypes that are both site and age specific. There was relative uniformity in the intestinal microflora during the breast-feeding period which was characterized by the dominancy of Lactobacillales species and proteobacteria, while Bacillales species were also common in the oral cavity. By day 21, however, Lactobacillales were dominant only in the oral and small intestinal microflora and Clostridiales and Bacteroidales predominated in the colon and cecum. The dominancy of Bacteroidales in the intestines of adult mice has been observed in previous studies (43, 48). The dramatic transition from a Lactobacillales/proteobacterium-rich microflora to a Clostridiales/Bacteroidetes-rich microflora in the colon after weaning suggests an important relationship between the environment and composition of the commensal bacteria. The proteobacteria and Lactobacillales species in the intestinal microflora are lactose-fermenting facultative anaerobic organisms that can grow under aerobic conditions and this is likely to explain their abundance during breast-feeding. In contrast, the standard diet of adult mice and humans is rich in fiber, which can be digested by Clostridiales and Bacteroidetes species, but not Lactobacillales and intestinal proteobacteria (29). This is also consistent our finding that the small intestinal microflora remains Lactobacillales/proteobacteria-rich after weaning, because unlike the colon, the small intestine is rich in monosaccharides and disaccharides. Thus, dietary changes during weaning are likely to be responsible for the major shift in the composition in the intestinal microflora. This is consistent with the fact that the presence of Clostridiales and Bacteroidetes species is modified by food composition (15, 48).
Closer inspection of the taxonomy within each order revealed significant variation in individual mice. For example, some mice had Leuconostoc citreum or Enterococcus faecalis, while others had Lactobacillus johnsonii and Lactococcus plantarum in the small intestine at day 3, although all of these bacteria are Lactobacillales species. Similarly, the microfloras of some mice contain Staphylococcus sciuri, while other mice had S. xylosus. Even though all mice had Clostridiales in the colon and cecum on day 21, there was significant species diversity in individual mice. The diversity in species may explain, at least in part, the differential innate stimulation of the microflora of individual mice at different stages of development.
A major finding of the present study was the identification of a developmental transition in TLR4-stimulatory activity which was associated with a shift from Gram-negative species such as E. coli that are strongly TLR4 stimulatory to Gram-positive Clostridiales and Gram-negative Bacteroidales that produce poorly stimulatory LPS. The TLR4-stimulatory activity of LPS is dependent on the number of phosphates and acyl chains in the lipid A moiety (19, 37). The known structures of LPS from all Bacteroidales species are non-TLR4 stimulatory (13, 35, 50), a finding consistent with previous studies (12, 24, 26). Therefore, the dominancy of Bacteroidales species and the disappearance of TLR4-stimulatory proteobacteria appears to explain the low ability of the cecal and colon microflora from 21-day-old mice to stimulate TLR4.
Although the colonic microflora has undetectable TLR4-stimulatory activity, the cecal and colonic microflora on day 21 elicited robust stimulatory responses in macrophages and mesothelial cells. This immunostimulatory activity was dependent on MyD88 but remained intact in primary cells from ASC-, RICK-, NOD1-, NOD2-, TLR2-, TLR4-, TLR5-, TLR9-, or TLR11-deficient mice. The finding that the microflora of the colon stimulates host cells via MyD88 is consistent with the observation that mutant mice deficient in MyD88 are highly susceptible to dextran sulfate sodium-induced epithelial damage in the adult colon (40). Potential bacterium-derived molecules responsible for the observed responses include single-stranded RNA (a TLR7/TLR8 ligand) that signal via MyD88 (20). Alternatively, the results could be explained by redundancy among different TLRs, although we did not find any dominant stimulatory activity as determined by biochemical fractionation (data not shown). In addition to stimulating via MyD88, the microflora of the 21-day-old intestine exhibited NOD1-stimulatory activity. The candidates for such an activity are DAP-type peptidoglycan containing bacteria, including Gram-negative beta-, gamma-, epsilonproteobacteria, Bacteroidetes, Gram-positive Clostridiales, Bacillaceae, and Paenibacillaceae of the Bacillales. The best candidates are two novel Porphyromonadaceae species, 21W05SI1 and 21W07-149, because we found a correlation between their presence and NOD1-stimulatory activity in the small intestine at day 21 (148 and 149 [numbers and rows in Fig. S2 and Table S2, respectively, in the supplemental material]). Because NOD1 regulates the susceptibility to allergic diseases (17, 49), it will be important to determine whether NOD1-stimulatory commensals affect development of allergic disease. This hypothesis can be now tested by manipulating the number of specific immunostimulatory bacteria through probiotics and/or prebiotics.
Our assay used BMM and mesothelial cells to estimate the immunostimulatory activities of TLR and NLR ligands in the present study. We recognize that in vivo innate stimulatory activity of the microbiota is much more complex due to differential ligand sensitivity of intestinal immune and epithelial cells and the accessibility of the immunostimulatory molecules, which is affected by physical barriers and active uptakes. Interestingly, our study demonstrated that NOD1 contributes via RICK to the stimulatory activity induced by adult small intestine and colonic microflora in the absence of MyD88. Intestinal epithelial cells are sensitive to TLR4 ligand on day 6 but become insensitive after weaning (day 28) (27). The latter may be important for switching the expression of antimicrobial peptides, which has been observed during mouse intestinal development (32). Importantly, mice and cells, such as BMM, are also reported to be desensitized to TLR stimulation after prolonged ligand exposure, and NLRs may play a role in immune signaling under conditions of TLR desensitization (22). Our study showed a dramatic increase in MyD88-dependent immunostimulatory activity in the intestine after weaning. Therefore, it will be interesting to test in future studies whether NOD1/RICK-mediated signaling mediates innate immune response in TLR-desensitized conditions, as well as the regulation of expression levels of PRRs.
The oral microflora possesses very low immunostimulatory activity. This could be explained by the relative low number of bacteria compared to the intestine and by the microflora composition. All tested Lactobacillales species possessed low NOD1-, NOD2-, and no TLR4-stimulatory activity (12). Similarly, Pseudomonas species which are found in the oral microflora had low or undetectable TLR4-stimulatory activity (12). Although the biological benefit of having the oral cavity colonized by bacteria with low immunostimulatory activity is unclear, it may important for enhancing the detection of oral pathogens by host cells or to minimize potentially harmful proinflammatory responses elicited by commensals.
The comparison between microflora of mice (the present study) and humans suggests both similarities and differences in bacterial populations between the two species. The dramatic increase in obligate anaerobes, that is, Clostridiales and Bacteroidales, in the intestinal microflora has been also reported in humans (28, 39). The Streptococcus group, a dominant bacteria in the human oral cavity (21, 38), is also dominant in the mouse oral cavity. However, we did not detect bifidobacteria, which are dominant bacteria in human infants (28, 39), as common bacteria in the mouse intestinal microflora. Furthermore, the dominancy of Staphylococcus species in the mouse was not found in the human oral microflora (21, 38). Moreover, even within the common bacterial orders, the actual families, and species were often different between mice and humans, which may have implications for the development and function of the immune system in mice versus humans.
Previous studies revealed that interactions between commensal bacteria and host through MyD88are important for epithelial cell homeostasis and IgA secretion in the intestine (40, 44). Furthermore, NOD1 and TLR9 were shown to play a role in the genesis of certain lymphoid tissues and the balance between regulatory T cells and IL-17- and IFN-γ-producing T cells in the intestine, respectively (5, 10). In these studies, the immunostimulatory activity was reconstituted with purified bacterial components (LPS, peptidoglycan fragment, and bacterial DNA) or whole bacteria (5, 10, 40). However, the amounts of introduced bacterial components including LPS and NOD1 ligand were ∼100-fold higher than the physiological levels found in weaned mice. In addition, bacterial species that were administered exogenously are different from those found in the intestine of adult mice. The effect of stimulation from commensals for Th17 differentiation through TLRs is still under debate (3, 18, 53). Thus, it will be important in future studies to take into account the physiological amounts of bacterial components and the particular species of commensal bacteria when interpreting data involving exogenous administration of bacteria and/or bacterium-derived components.
The role of host immune system such as IgA in the development of the microflora remains controversial (25, 42, 44). The microflora of NOD1-deficient mice was reported to contain higher numbers of Clostridiales in the ileal biofilm (5), and MyD88-deficient mice contained increased percentages of particular bacteria including Lactobacillaceae and Porphyromonadaceae species (51). However, it remains unclear how these bacterial populations stimulate host signaling via PRRs or the mechanism by which PRRs regulate commensal bacteria. Clearly, additional studies are needed to understand how host immunity controls the microflora and conversely, the mechanism whereby the bacterial microflora affects host immunity.
Supplementary Material
Acknowledgments
We thank T. Koseki (Tohoku University) for stimulating discussions and L. Franchi, Y.-G. Kim, J.-W. Park, S. Koonse, T. Eigenbrod, T. Standiford, and K. Eaton (University of Michigan) for materials.
The study was supported by NIH grants R01DE018503 (to N.I.) and R01 DK61707 (to G.N.). This study is also supported in part by the Global COE program “Center for Practical Chemical Wisdom” by MEXT (to S.T.).
The authors have no conflicting financial interests.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 23 November 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Reference deleted.
- 2.Artis, D. 2008. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat. Rev. Immunol. 8:411-420. [DOI] [PubMed] [Google Scholar]
- 3.Atarashi, K., J. Nishimura, T. Shima, Y. Umesaki, M. Yamamoto, M. Onoue, H. Yagita, N. Ishii, R. Evans, K. Honda, and K. Takeda. 2008. ATP drives lamina propriaT(H)17 cell differentiation. Nature 455:808-812. [DOI] [PubMed] [Google Scholar]
- 4.Bartosch, S., A. Fite, G. T. Macfarlane, and M. E. McMurdo. 2004. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl. Environ. Microbiol. 70:3575-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouskra, D., C. Brézillon, M. Bérard, C. Werts, R. Varona, I. G. Boneca, and G. Eberl. 2008. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456:507-510. [DOI] [PubMed] [Google Scholar]
- 6.Chen, G. Y., M. H. Shaw, G. Redondo, and G. Núñez. 2008. The innate immune receptor NOD1 protects the intestine from inflammation-induced tumorigenesis. Cancer Res. 68:10060-10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fritz, J. H., and S. E. Girardin. 2005. How Toll-like receptors and Nod-like receptors contribute to innate immunity in mammals. J. Endotoxin Res. 11:390-394. [DOI] [PubMed] [Google Scholar]
- 9.Fukata, M., A. Chen, A. S. Vamadevan, J. Cohen, K. Breglio, S. Krishnareddy, D. Hsu, R. Xu, N. Harpaz, A. J. Dannenberg, K. Subbaramaiah, H. S. Cooper, S. H. Itzkowitz, and M. T. Abreu. 2007. Toll-like receptor-4 promotes the development of colitis-associated colorectal tumors. Gastroenterology 133:1869-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall, J. A., N. Bouladoux, C. M. Sun, E. A. Wohlfert, R. B. Blank, Q. Zhu, M. E. Grigg, J. A. Berzofsky, and Y. Belkaid. 2008. Commensal DNA limits regulatory T-cell conversion and is a natural adjuvant of intestinal immune responses. Immunity 29:637-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hampe, J., A. Cuthbert, P. J. Croucher, M. M. Mirza, S. Mascheretti, S. Fisher, H. Frenzel, K. King, A. Hasselmeyer, A. J. MacPherson, S. Bridger, van S. Deventer, A. Forbes, S. Nikolaus, J. E. Lennard-Jones, U. R. Foelsch, M. Krawczak, C. Lewis, S. Schreiber, and C. G. Mathew. 2001. Association between insertion mutation in NOD2 gene and Crohn's disease in German and British populations. Lancet 357:1925-1928. [DOI] [PubMed] [Google Scholar]
- 12.Hasegawa, M., K. Yang, M. Hashimoto, J. H. Park, Y. Fujimoto, G. Núñez, K. Fukase, and N. Inohara. 2006. Differential release and distribution of NOD1 and NOD2 immunostimulatory molecules among bacterial species and environments. J. Biol. Chem. 281:29054-29063. [DOI] [PubMed] [Google Scholar]
- 13.Hashimoto, M., F. Kirikae, T. Dohi, S. Adachi, S. Kusumoto, Y. Suda, T. Fujita, H. Naoki, and T. Kirikae. 2002. Structural study on lipid A and the O-specific polysaccharide of the lipopolysaccharide from a clinical isolate of Bacteroides vulgatus from a patient with Crohn's disease. Eur. J. Biochem. 269:3715-3721. [DOI] [PubMed] [Google Scholar]
- 14.Holler, E., G. Rogler, H. Herfarth, J. Brenmoehl, P. J. Wild, J. Hahn, G. Eissner, J. Schölmerich, and R. Andreesen. 2004. Both donor and recipient NOD2/CARD15 mutations associate with transplant-related mortality and GvHD following allogeneic stem cell transplantation. Blood 104:889-894. [DOI] [PubMed] [Google Scholar]
- 15.Hooper, L. V., T. Midtvedt, and J. I. Gordon. 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22:283-307. [DOI] [PubMed] [Google Scholar]
- 16.Hugot, J. P., M. Chamaillard, H. Zouali, S. Lesage, J. P. Cézard, J. Belaiche, S. Almer, C. Tysk, C. A. O'Morain, M. Gassull, V. Binder, Y. Finkel, A. Cortot, R. Modigliani, P. Laurent-Puig, C. Gower-Rousseau, J. Macry, J. F. Colombel, M. Sahbatou, and G. Thomas. 2001. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411:599-603. [DOI] [PubMed] [Google Scholar]
- 17.Hysi, P., M. Kabesch, M. F. Moffatt, M. Schedel, D. Carr, Y. Zhang, B. Boardman, von E. Mutius, S. K. Weiland, W. Leupold, C. Fritzsch, N. Klopp, A. W. Musk, A. James, G. Núñez, N. Inohara, and W. O. Cookson. 2005. NOD1 variation, immunoglobulin E and asthma. Hum. Mol. Genet. 14:935-941. [DOI] [PubMed] [Google Scholar]
- 18.Ivanov, I. I., R. de L. Frutos, N. Manel, K. Yoshinaga, D. B. Rifkin, R. B. Sartor, B. B. Finlay, and D. R. Littman. 2008. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4:337-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin, M. S., and J. O. Lee. 2008. Structures of the Toll-like receptor family and its ligand complexes. Immunity 29:182-191. [DOI] [PubMed] [Google Scholar]
- 20.Kawai, T., and S. Akira. 2009. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 21:317-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keijser, B. J., E. Zaura, S. M. Huse, J. M. van der Vossen, F. H. Schuren, R. C. Montijn, J. M. ten Cate, and W. Crielaard. 2008. Pyrosequencing analysis of the oral microflora of healthy adults. J. Dent. Res. 87:1016-1020. [DOI] [PubMed] [Google Scholar]
- 22.Kim, Y. G., J. H. Park, M. H. Shaw, L. Franchi, N. Inohara, and G. Núñez. 2008. The cytosolic sensors Nod1 and Nod2 are critical for bacterial recognition and host defense after exposure to Toll-like receptor ligands. Immunity 28:246-257. [DOI] [PubMed] [Google Scholar]
- 23.Ley, R. E., F. Bäckhed, P. Turnbaugh, C. A. Lozupone, R. D. Knight, and J. I. Gordon. 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U. S. A. 102:11070-11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindberg, A. A., A. Weintraub, U. Zähringer, and E. T. Rietschel. 1990. Structure-activity relationships in lipopolysaccharides of Bacteroides fragilis. Rev. Infect. Dis. 12(Suppl. 2):S133-S141. [DOI] [PubMed] [Google Scholar]
- 25.Loh, G., F. Brodziak, and M. Blaut. 2008. The Toll-like receptors TLR2 and TLR4 do not affect the intestinal microbiota composition in mice. Environ. Microbiol. 10:709-715. [DOI] [PubMed] [Google Scholar]
- 26.Lorenz, E., D. D. Patel, T. Hartung, and D. A. Schwartz. 2002. Toll-like receptor 4 (TLR4)-deficient murine macrophage cell line as an in vitro assay system to show TLR4-independent signaling of Bacteroides fragilis lipopolysaccharide. Infect. Immun. 70:4892-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lotz, M., D. Gütle, S. Walther, S. Ménard, C. Bogdan, and M. W. Hornef. 2006. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J. Exp. Med. 203:973-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackie, R. I., A. Sghir, and H. R. Gaskins. 1999. Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 69:1035S-1045S. [DOI] [PubMed] [Google Scholar]
- 29.Mahowald, M. A., F. E. Rey, H. Seedorf, P. J. Turnbaugh, R. S. Fulton, A. Wollam, N. Shah, C. Wang, V. Magrini, R. K. Wilson, B. L. Cantarel, P. M. Coutinho, B. Henrissat, L. W. Crock, A. Russell, N. C. Verberkmoes, R. L. Hettich, and J. I. Gordon. 2009. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. U. S. A. 106:5859-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masumoto, J., K. Yang, S. Varambally, M. Hasegawa, S. A. Tomlins, S. Qiu, Y. Fujimoto, A. Kawasaki, S. J. Foster, Y. Horie, T. W. Mak, G. Núñez, A. M. Chinnaiyan, K. Fukase, and N. Inohara. 2006. NOD1 acts as an intracellular receptor to stimulate chemokine production and neutrophil recruitment in vivo. J. Exp. Med. 203:203-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGovern, D. P., P. Hysi, T. Ahmad, van D. A. Heel, M. F. Moffatt, A. Carey, W. O. Cookson, and D. P. Jewell. 2005. Association between a complex insertion/deletion polymorphism in NOD1 (CARD4) and susceptibility to inflammatory bowel disease. Hum. Mol. Genet. 14:1245-1250. [DOI] [PubMed] [Google Scholar]
- 32.Ménard, S., V. Förster, M. Lotz, D. Gütle, C. U. Duerr, R. L. Gallo, B. Henriques-Normark, K. Pütsep, M. Andersson, E. O. Glocker, and M. W. Hornef. 2008. Developmental switch of intestinal antimicrobial peptide expression. J. Exp. Med. 205:183-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morelli, L. 2008. Postnatal development of intestinal microflora as influenced by infant nutrition. J. Nutr. 138:1791S-1795S. [DOI] [PubMed] [Google Scholar]
- 34.Muyzer, G., de E. C. Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogawa, T. 1993. Chemical structure of lipid A from Porphyromonas (Bacteroides) gingivalis lipopolysaccharide. FEBS Lett. 332:197-201. [DOI] [PubMed] [Google Scholar]
- 36.Ogura, Y., D. K. Bonen, N. Inohara, D. L. Nicolae, F. Chen, R. Ramos, H. Britton, T. Moran, R. Karaliuskas, R. H. Duerr, J. P. Achkar, S. R. Brant, T. M. Bayless, B. S. Kirschner, S. B. Hanauer, G. Núñez, and J. H. Cho. 2001. Susceptibility to Crohn's disease by a frameshift mutation in NOD2, resulting in lipopolysaccharide hyporesponsiveness. Nature 411:603-606. [DOI] [PubMed] [Google Scholar]
- 37.Park, B. S., D. H. Song, H. M. Kim, B. S. Choi, H. Lee, and J. O. Lee. 2009. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458:1191-1195. [DOI] [PubMed] [Google Scholar]
- 38.Paster, B. J., I. Olsen, J. A. Aas, and F. E. Dewhirst. 2006. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol. 2000 42:80-87. [DOI] [PubMed] [Google Scholar]
- 39.Picard, C., J. Fioramonti, A. Francois, T. Robinson, F. Neant, and C. Matuchansky. 2005. Review article: bifidobacteria as probiotic agents-physiological effects and clinical benefits. Aliment Pharmacol. Ther. 22:495-512. [DOI] [PubMed] [Google Scholar]
- 40.Rakoff-Nahoum, S., J. Paglino, F. Eslami-Varzaneh, S. Edberg, and R. Medzhitov. 2004. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118:229-241. [DOI] [PubMed] [Google Scholar]
- 41.Round, J. L., and S. K. Mazmanian. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9:313-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sait, L., M. Galic, R. A. Strugnell, and P. H. Janssen. 2003. Secretory antibodies do not affect the composition of the bacterial microbiota in the terminal ileum of 10-week-old mice. Appl. Environ. Microbiol. 69:2100-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salzman, N. H., H. de Jong, Y. Paterson, H. J. Harmsen, G. W. Welling, and N. A. Bos. 2002. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology 148:3651-3660. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki, K., and S. Fagarasan. 2008. How host-bacterial interactions lead to IgA synthesis in the gut. Trends Immunol. 29:523-531. [DOI] [PubMed] [Google Scholar]
- 45.Takeuchi, O., K. Hoshino, and S. Akira. 2000. TLR2-deficient and MyD88-deficient mice are highly susceptible to Staphylococcus aureus infection. J. Immunol. 165:5392-5396. [DOI] [PubMed] [Google Scholar]
- 46.Tanamoto, K., U. Zähringer, G. R. McKenzie, C. Galanos, E. T. Rietschel, O. Lüderitz, S. Kusumoto, and T. Shiba. 1984. Biological activities of synthetic lipid A analogs: pyrogenicity, lethal toxicity, anticomplement activity, and induction of gelation of Limulus amebocyte lysate. Infect. Immun. 44:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teng, Y. T. 2006. Protective and destructive immunity in the periodontium. 1. Innate and humoral immunity and the periodontium. J. Dent. Res. 85:198-208. [DOI] [PubMed] [Google Scholar]
- 48.Turnbaugh, P. J., F. Bäckhed, L. Fulton, and J. I. Gordon. 2008. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3:213-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weidinger, S., N. Klopp, L. Rummler, S. Wagenpfeil, N. Novak, H. J. Baurecht, W. Groer, U. Darsow, J. Heinrich, A. Gauger, T. Schafer, T. Jakob, H. Behrendt, H. E. Wichmann, J. Ring, and T. Illig. 2005. Association of NOD1 polymorphisms with atopic eczema and related phenotypes. J. Allergy Clin. Immunol. 116:177-184. [DOI] [PubMed] [Google Scholar]
- 50.Weintraub, A., U. Zähringer, H. W. Wollenweber, U. Seydel, and E. T. Rietschel. 1989. Structural characterization of the lipid A component of Bacteroides fragilis strain NCTC 9343 lipopolysaccharide. Eur. J. Biochem. 183:425-431. [DOI] [PubMed] [Google Scholar]
- 51.Wen, L., R. E. Ley, P. Y. Volchkov, P. B. Stranges, L. Avanesyan, A. C. Stonebraker, C. Hu, F. S. Wong, G. L. Szot, J. A. Bluestone, J. I. Gordon, and A. V. Chervonsky. 2008. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455:1109-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamamoto, M., S. Sato, H. Hemmi, K. Hoshino, T. Kaisho, H. Sanjo, O. Takeuchi, M. Sugiyama, M. Okabe, K. Takeda, and S. Akira. 2003. Role of adaptor TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science 301:640-643. [DOI] [PubMed] [Google Scholar]
- 53.Zaph, C., Y. Du, S. A. Saenz, M. G. Nair, J. G. Perrigoue, B. C. Taylor, A. E. Troy, D. E. Kobuley, R. A. Kastelein, D. J. Cua, Y. Yu, and D. Artis. 2008. Commensal-dependent expression of IL-25 regulates the IL-23-IL-17 axis in the intestine. J. Exp. Med. 205:2191-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.