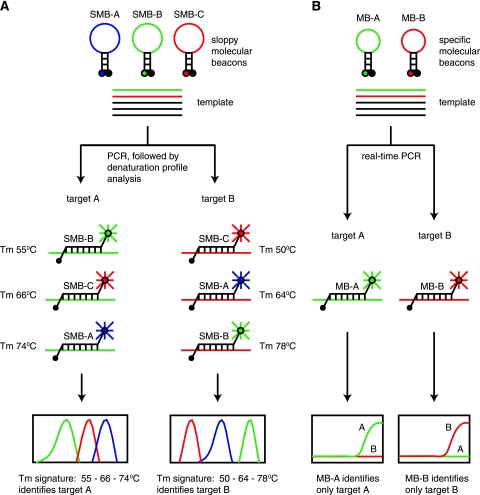
FIG. 1.
Two paradigms of target identification. (A) Sloppy molecular beacon approach. Three sloppy molecular beacons with different semispecific probe regions are placed in a PCR with an unknown target. Each probe is distinguished by a unique fluorophore. Asymmetrical PCR is performed, and the Tm of each probe target-hybrid is measured. Each probe generates a Tm that is specific for a limited number of targets such that probe A, probe B, and probe C generate Tm values of 55°C, 66°C, and 74°C, respectively, with target A and Tm values of 64°C, 78°C, and 50°C, respectively, with target B. All three Tm values are combined into a single species-specific signature that identifies the unknown target. Hundreds of different targets can theoretically be identified and distinguished with the same three probes in this manner. (B) Conventional approach. Two molecular beacons with probe regions that are specific for a single target sequence are placed in a PCR with an unknown target. Each molecular beacon is distinguished by a unique fluorophore or reaction well. PCR is performed, and the presence or absence of each molecular beacon signal is assessed. The unknown target is identified by determining which of the molecular beacons in a reaction produces a positive signal. The number of different target sequences that can be identified is limited to the number of different molecular beacons that are used in the assay.