FIG. 9.
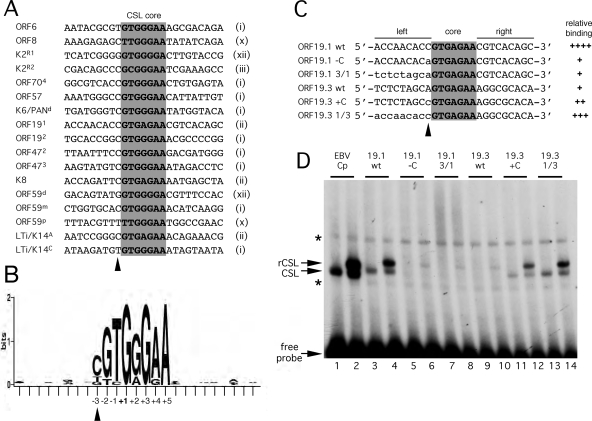
Defining a signature for CSL binding sites in the KSHV genome. (A) Alignment of 17 confirmed CSL binding sites from the KSHV genome. The core sequence is shaded, and the core motif type, as defined in Fig. 1A, is noted on the right. Labeling of individual sites from K2/vIL6, K6/PAN, ORF59, and LTi/K14 follows published naming schemes (4, 33, 34, 36). (B) A positional nucleotide frequency pattern (sequence logo) for the sites listed in panel A was generated using WebLogo (9). Only positions +1, +3, and +5 are invariant. This visual representation emphasizes the strong preference for a cytosine at position −3 and the striking absence of sequence conservation beyond the eight central positions (−3 to +5). (C) Sequences of probes used to test the contributions of flanking residues to the differential binding of CSL to ORF19 sites 1 and 3. The wild-type sequence is shown in uppercase, and the altered sequence is shown in lowercase. (D) Gel mobility shift analysis of the probes shown in panel C. Each was labeled to an identical specific activity and then incubated with HeLa whole-cell extract from mock-transfected cells (odd-numbered lanes) or cells expressing tagged CSL (even-numbered lanes). The EBV Cp site (lanes 1 and 2) was included as a reference. Positions of the unbound probe and complexes corresponding to endogenous (CSL) or transfected (rCSL) CSL are indicated. Nonspecific complexes are marked with asterisks. The ability of each probe to form a stable complex with CSL is summarized in panel C.