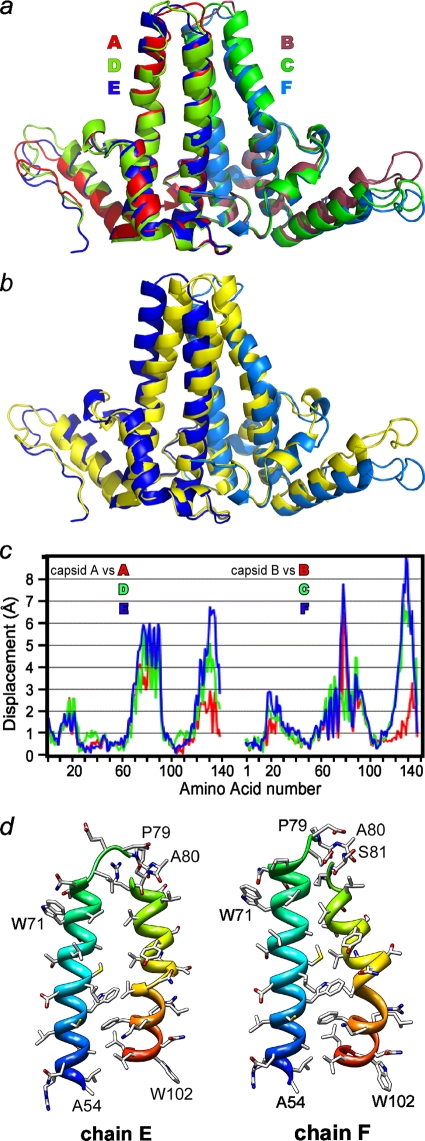
FIG. 2.
Overlays of dimers show asymmetry and divergence from capsid structure viewed tangent to the virus surface. (a) Cp149-Y132A dimers are asymmetric and structurally unique. The monomers fall into two structural classes based on the conformation of the long helices at the intradimer interface. Even so, obvious differences at the tip of the spike (top center of the figure) and at the turn at the end of the interdimer contacts are apparent. (b) Overlay of the EF Cp149-Y132A dimer (with the E and F subunits colored dark blue and marine blue, as in panel a) and dimer from capsid (AB from the PDB no. 1QGT structure [48], in yellow) to allow comparison of free and dimer structures. There are substantial differences between the two conformations at the periphery, even though central cores, or chasses, are identical. Unlike Cp149-Y132A, the dimer from capsid has well-maintained twofold symmetry (see the supplemental material). (c) For quantitative analysis, as well for all overlays in this figure, chassis α carbons (corresponding to residues 1 to 10, 26 to 62, and 95 to 110) of A, D, and E subunits of Cp149-Y132A were aligned with equivalent residues from a capsid A subunit; distances between equivalent α carbons throughout the dimers are reported. Though not specifically constrained, both half dimers show the same pattern of similarity and difference. (d) A side-by-side comparison of the E (left) and F (right) halves of the four-helix bundle spike shows the asymmetry of the Cp149-Y132A dimers. This view shows the complementary surfaces that are buried at the dimer interface. The segments from A54 to W71 have approximately the same alignments to facilitate comparison. Note that residues 79 and 80 are part of the random coil turn in chain F.