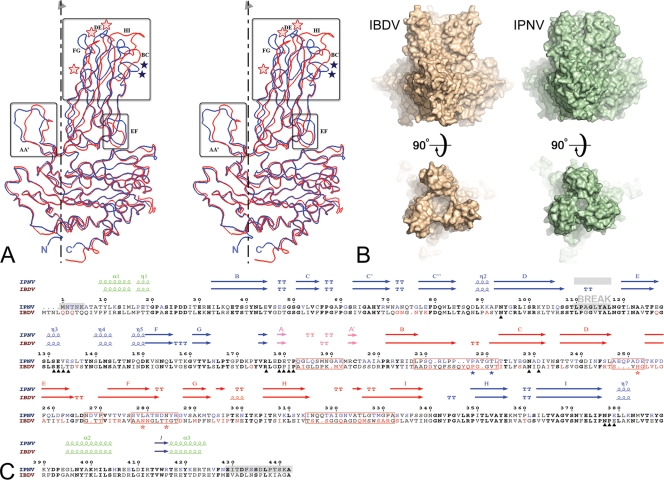
FIG. 2.
Structural comparison between IPNV and IBDV VP2. (A) Structural alignment of IPNV VP2 and IBDV VP2 subunits. The VP2 protomers were superimposed on their respective S domains and are represented as blue (IPNV) and red (IBDV) Cα traces. The molecular 3-fold axis, vertical in the figure, is represented by a broken line. Regions of structural divergence are boxed. Blue and red stars denote residues implicated in virulence of IPNV (positions 217 and 221) and IBDV (positions 253, 279, and 284), respectively. (B) Morphological differences of IPNV VP2 and IBDV VP2 spikes. The molecular surfaces of the respective VP2 trimers are shown in orthogonal views through the surface (side views at top) and from the outside (top views at bottom) of the particle. Note the more compact arrangement observed for IPNV VP2, with a narrower opening at the center of the trimer and shorter projections at the top of the spike. (C) Amino acid sequence alignment of IPNV and IBDV VP2s with their respective secondary structure elements colored according to domains as described in the legend of Fig. 1C (green, base; blue, shell; orange, projection; pink, AA′ flap). Residues in bold font are not only conserved across IPNV and IBDV sequences but are also strictly conserved within the respective genera. Residues shown in blue and red are variable between serotypes of IPNV and IBDV, respectively. Red boxes represent regions of structural dissimilarities between IPNV and IBDV also highlighted in panel A. A gray background indicates regions that were disordered in the crystal structure (including the break in density in domain S). Black triangles under the sequences mark the amino acids lining the conserved groove at the base of the spike described in the text and in Fig. 3. Blue and red stars under the sequences denote residues implicated in virulence of IPNV and IBDV, respectively, as in panel A. Note that position 231, marked with a triangle, is not in bold font because it is not aspartic acid in one of the available IPNV sequences, whereas it is aspartic in every other birnavirus that has been sequenced to date. Since this is the conserved integrin binding motif, it is possible that the particular sequence has an error at this position.