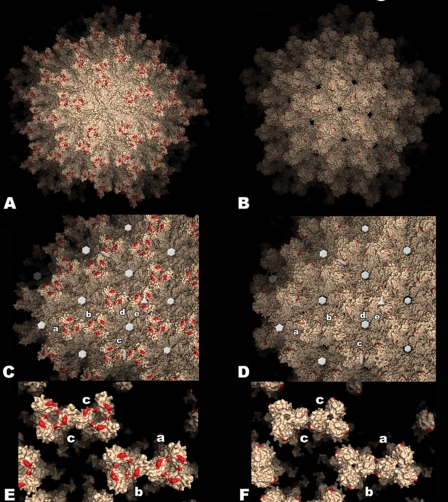
FIG. 4.
Model of the IPNV virus particle. Surface representation of the T=13l lattice of IBDV (left panels) and IPNV (right panels) virions. Red patches indicate residues implicated in cell culture adaptation and virulence in IBDV (His253, Asn279, and Thr284) and IPNV (Pro217 and Thr221). (A and B) View along a 5-fold axis of the particle. (C and D) View along a 3-fold axis. Five icosahedrally independent VP2 trimers are labeled a to e according to the nomenclature established for the IBDV virion (5). Twofold, 3-fold, 5-fold, and pseudo-6-fold axes are indicated with ellipses, triangles, pentagons, and hexagons, respectively (the holes on these axes in the IPNV virion result from the disordered loop in this region, as explained in the text). (E and F) Close-up view of contacts between spikes involving domain P. Note the weaker contacts between spikes a-b and c-c, as well as the more closed organization of the spike head in the IPNV virion.