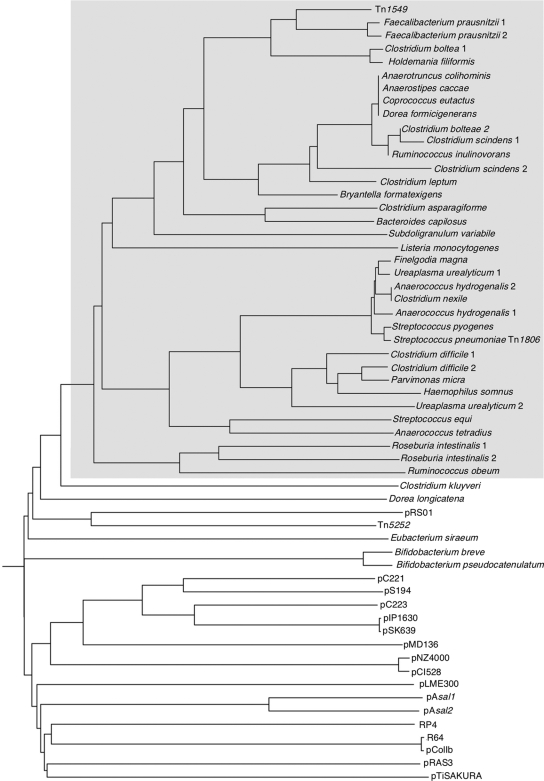
FIG. 6.
Phylogenetic relationship among proteins belonging to the 3H family of MOBp-superfamily relaxases. The tree was constructed from sequences available in data banks by use of the neighbor-joining method. The origin of relaxase-containing plasmids, transposons, or the bacterial host is indicated at each branch. The phyletic group including RlxTn1549 is gray shadowed. Accession numbers of relaxases: Tn1549, AAF72355; A. hydrogenalis 1, ZP_03303665.1; A. hydrogenalis 2, ZP_03303763.1; A. tetradius, NC_ACGC01000033.1; A. caccae, ZP_02419708.1; A. colihominis, NZ_ABGD02000024.1; B. capillosus, ZP_02036891; B. breve, ZP_03619377; B. pseudocatenulatum, NZ_ABXX02000003; B. formatexigens, ZP_03686097; C. asparagiforme DSM 15981, NZ_ACCJ01000017.1; C. bolteae, ZP_02085866.1 and ZP_02085146.1; C. difficile, YP_001088377.1 and YP_001086893.1; C. kluyveri, YP_0013933676.1; C. leptum, ZP_02081957.1; C. nexile, ZP_03288837.1; C. scindens, ZP_02429884.1 and ZP_02432986.1; C. eutactus, ZP_02207623.1; D. formicigenerans, ZP_02235231.1; D. longicatena, ZP_01994204.1; E. siraeum, ZP_02422210.1; F. prausnitzii, ZP_02093051.1 and ZP_02090960.1; F. magna, YP_001692251.1; H. somnus, YP_001784516.1; H. filiformis, ZP_03636025.1; YP_0011784516.1; L. monocytogenes, ZP_00229823.1; P. micra, ZP_02093865.1; R. intestinalis, ZP_03483057.1 and ZP_03485669.1; R. inulinovorans, ZP_03752747.1; R. obeum, ZP-01964421.1 and ZP_01962871.1; S. equi, CAP20357; S. pneumoniae AP200 (Tn1806), EF469826; S. pyogenes, CAQ56294.1; S. variabile, ZP_03776736.1; U. urealyticum serovar 9, ZP_03079552.1; U. urealyticum serovar 13, ZP_02696018.1; ColIbP-9, NP_052501.1; pAH82, NP_862611.1; pAsal1, CAD48422.1; pAsal2, NP_710170.1; pC221, CAA26106; pC223, NP_943089; pCI1528, AAB28188.1; pFN1, NP_862674.1; pIP1629, AAD02378.1; pIP1630, AAD02405.1; pLME300, CAD59910; pMD136, NP_037562.1; pNZ4000, NP_053037; pRAS3, NP_387457.1; pRS01, AAB06502.1; pS194, NP_976272; pSK639, NP_976278; RP4, Q00191; R64, B38529; Tn5252, NP_358551.