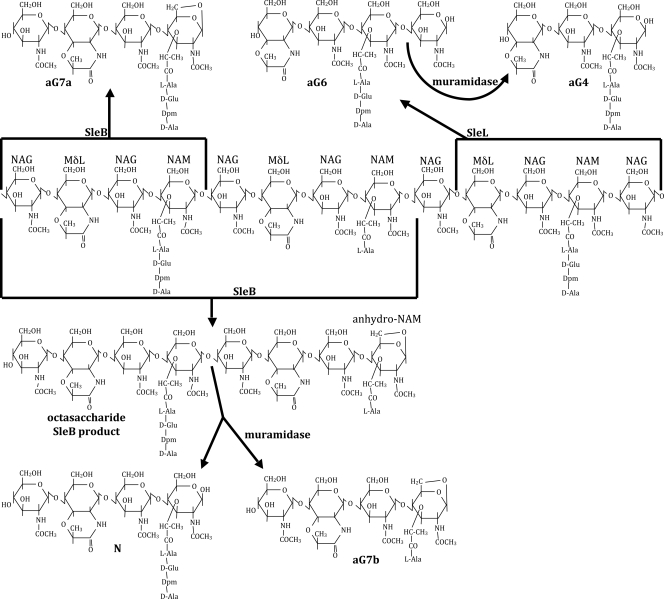
FIG. 1.
Spore PG structure and hydrolysis. The central structure shows a representative spore PG strand with alternating NAG and NAM or muramic-δ-lactam (MδL) residues and with tetrapeptide or l-Ala side chains on the NAM residues. Forked arrows originate at sites of hydrolysis by the indicated enzymes and point to muropeptide products. The indicated “aG” muropeptide names are as previously published (7, 11). SleB lytic transglycosylase activity produces muropeptides terminating in anhydro-NAM. Cleavage at adjacent NAM residues produces the tetrasaccharide aG7a or aG7b, while cleavage further apart can produce octasaccharides or larger fragments. These can be further cleaved by muramidase treatment, resulting in the production of tetrasaccharide N, which terminates in NAM. The N-acetylglucosaminidase activity of SleL produces tetrasaccharides terminating in NAG, which can be further cleaved by muramidase to trisaccharides terminating in NAM.