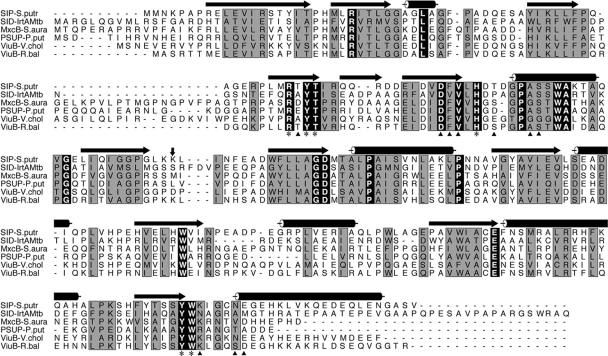
FIG. 3.
Sequence analysis of IrtA-NTD. A sequence alignment of the M. tuberculosis IrtA-NTD with other putative iron-siderophore utilization proteins is shown. The sequences were aligned by using CLUSTAL W (25), and the figure was generated with ALSCRIPT (1). Terms: SIP-S.Putr, siderophore-interacting protein from Shewanella putrefaciens; SID-IrtAMtb, IrtA-NTD from M. tuberculosis; MxcB-S.aura, myxochelin iron-binding protein from Stigmatella aurantiaca; PSUP-P.put, putative iron-chelator utilization protein from Pseudomonas putida; ViuB-V.chol, vibriobactin utilization protein from V. cholerae; ViuB-R.bal, vulnibactin utilization protein from Rhodopirellula baltica. The secondary structural elements at the top of the sequences are from the crystal structure of SIP-S.putr (PDB ID 2GPJ). Cylinders represent α-helices, and arrows denote β-strands. Residues labeled with an asterisk at the bottom of the sequences are those in the SIP-S.putr structure whose side chains interact with the bound FAD. These residues are identical among the aligned sequences. Residues marked with a triangle are those having direct hydrogen bonds from main chain atoms to the bound FAD. A vertical arrow marks the domain boundary between the N- and C-terminal domains. The C-terminal domain of these proteins has a β1α1β2α2β3α3 Rossmann fold, α4 connecting β3 and β4, and a second Rossmann fold lacking the sixth strand as is observed in other FAD-binding proteins (4).