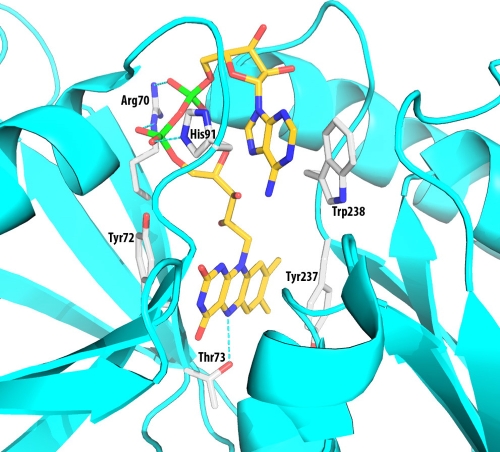
FIG. 8.
Model structure showing the FAD-binding pocket IrtA-NTD. The model was constructed based on sequence alignments and the structure of the SUP from S. putrefaciens (PDB entry 2GPJ). Only side chains that are directly involved in the binding interactions are shown. These side chains are conserved in most of the SUP sequences. There are also many H-bonds and pi-electron interactions with the protein main chain, which are not shown in the figure for clarity. The residue numbers are from the IrtA protein sequence. The figure was generated by using the program PYMOL (http://www.pymol.org).