Summary
Background
Piwi-interacting RNAs (piRNAs) are ~24–30 nucleotide regulatory RNAs that are abundant in animal gonads and early embryos. The best characterized piRNAs mediate a conserved pathway that restricts transposable elements, and these frequently engage a "ping-pong" amplification loop. Certain stages of mammalian testis also accumulate abundant piRNAs of unknown function, which derive from non-coding RNAs that are depleted in TE content and do not engage in ping-pong.
Results
We report that the 3' untranslated regions (3' UTRs) of an extensive set of messenger RNAs (mRNAs) are processed into piRNAs in Drosophila ovaries, murine testes, and Xenopus eggs. Analysis of small RNA data from different mutants and Piwi-class immunoprecipitates indicates that their biogenesis depends on primary piRNA components but not ping-pong components. Several observations suggest that mRNAs are actively selected for piRNA production. First, genic piRNAs do not accumulate in proportion to the level of their host transcripts, and many highly expressed transcripts lack piRNAs. Second, piRNA-producing mRNAs in Drosophila and mouse are enriched for specific gene ontology categories distinct from those of simply abundant transcripts. Third, the levels of Traffic Jam, whose 3' UTR generates abundant piRNAs, are increased in piwi mutant follicle clones. These data suggest that selection of cellular transcripts by the primary piRNA pathway is not fortuitous, but instead an active process with regulatory consequences.
Conclusions
Our work reveals a conserved primary piRNA pathway that selects and metabolizes the 3' UTRs of a broad set of cellular transcripts, providing insights into piRNA biogenesis and function. These data strongly increase the breadth of Argonaute-mediated small RNA systems in metazoans.
Introduction
The Piwi family comprises a subclass of Argonaute proteins that are primarily expressed in invertebrate and vertebrate gonads, and bind ~24–30 nucleotide RNAs termed Piwi-interacting RNAs (piRNAs) [1]. The best-studied function of the piRNA pathway is to maintain genomic integrity in the germline by restricting transposable element (TE) transcripts. Accordingly, most previously described Drosophila piRNAs derive from active TEs or from piRNA "master loci", which are rich in defective TE sequences and often transcribed in antisense orientation. These piRNAs fuel an amplification cycle that selectively amplifies a post-transcriptional defense against active TEs, termed "piRNA ping-pong" [2, 3]. This pathway is ancient and deeply conserved amongst metazoans[4–6].
Much less is known about piRNAs that do not derive from TEs or other repetitive sequences. For example, intergenic non-coding transcripts that are relatively depleted in TEs generate extremely abundant piRNAs in mammalian pachytene testes [7–10]. The biogenesis and function of pachytene piRNAs are largely mysterious. Their organization is analogous in some respects to Drosophila piRNA master clusters; however, their lack of obvious antisense counterparts has made it difficult to infer their regulatory targets. It is presumed that pachytene piRNAs are made by a primary piRNA processing pathway that does not involve amplification.
We recently analyzed Drosophila OSS cells [11], a somatic ovarian cell line inferred to derive from a prefollicular cell progenitor [12]. In that study, we demonstrated that OSS cells expresses Piwi but not Aub or AGO3, and consequently express only primary piRNAs [11]. This cell system is simplified relative to the intact gonad, which is composed of diverse cell types with varied expression of Piwi-class proteins. While the majority of OSS cell piRNAs correspond to TEs and major piRNA clusters, we describe here a second substantial population of piRNAs derived from protein-coding genes. Genic piRNAs preferentially arise from 3’ untranslated regions (3' UTRs) and are produced by a primary piRNA pathway that does not require ping-pong components. Largescale piRNA sequencing in vertebrate cells reveals that the 3' UTR-directed piRNA pathway is conserved in vertebrates such as mice and Xenopus.
A shared feature of mRNA populations metabolized by this pathway in different species is that piRNA accumulation correlates only modestly with transcript abundance; a substantial proportion of very highly expressed mRNAs do not generate abundant piRNAs. This suggests that this pathway does not sample cellular transcripts indiscriminately. In support of this, gene ontology (GO) terms enriched amongst transcripts with abundant piRNAs were distinct from those of highly expressed mRNAs, and many GO terms were shared by piRNA-producing transcripts in fly and mouse gonads. In summary, we find that a substantial and conserved primary piRNA biogenesis pathway acts selectively on the 3' UTRs of messenger RNAs that function in gonadal and germline development.
Results
Distinct siRNA and piRNA pathways act upon 3' UTRs in Drosophila OSS cells
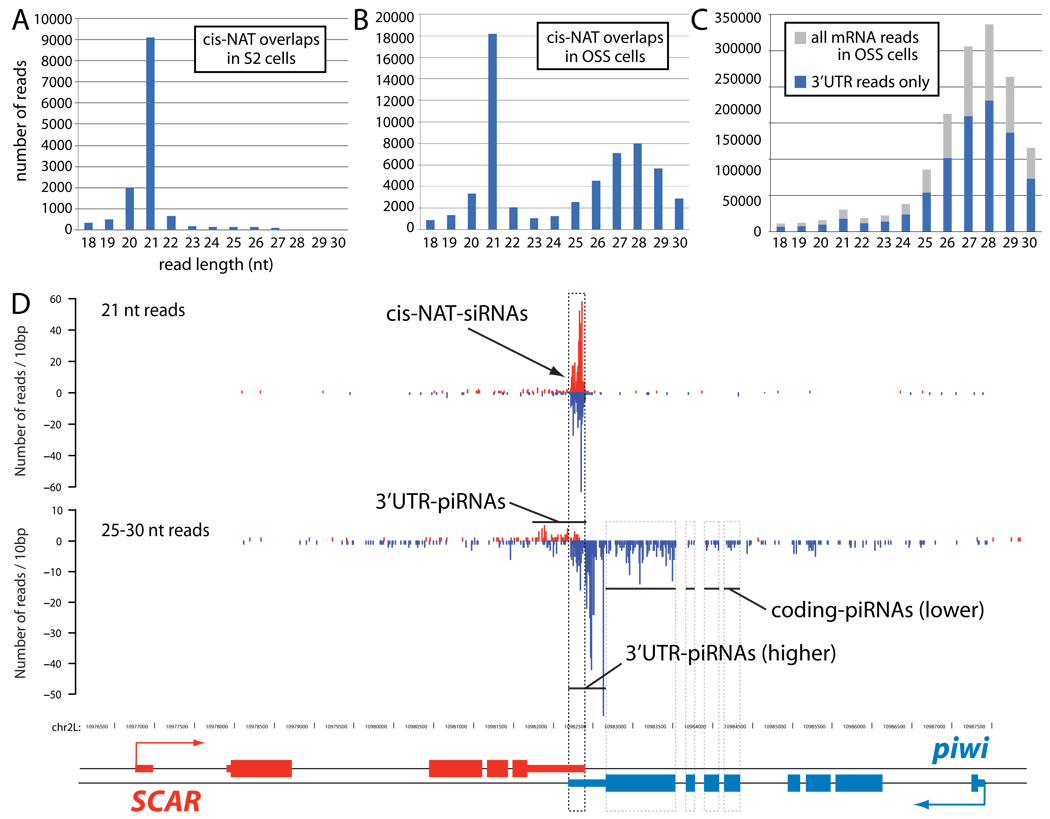
We recently showed that the Drosophila OSS cell line not only expresses miRNAs, but generates abundant piRNAs and siRNAs from transposable elements (TEs) [11]. Its accumulation of abundant piRNAs is shared with gonadal tissues and early embryos [13–16]. On the other hand, most Drosophila cell types are capable of mounting an RNAi response, either to endogenous or exogenous triggers [17–19]. For example, somatic and germline cells generate endo-siRNAs from TEs and from the overlap regions of convergently transcribed protein-coding genes ("3' cis-NAT-siRNAs") [20–23]. Indeed, we categorized >18,000 cis-NAT-siRNAs from OSS small RNA reads, of which 75% were accounted for by the overlap regions of 86 3' cis-NAT pairs that each generated >50 siRNAs (Figure 1A and Supplementary Table S1A).
Figure 1.
Distinct spatial patterns of siRNAs and piRNAs mapped to mRNAs. (A) 3' cis-natural antisense transcript (cis-NAT) overlap regions generate nearly exclusively 21 nt reads (siRNAs) in S2 cells. (B) cis-NAT overlaps generate both siRNAs and piRNAs in OSS cells. (C) Analysis of all mRNA-derived reads (blue) and 3' UTR reads (grey) from OSS cells shows that piRNAs are preferentially produced from 3' UTRs. (D) Small RNAs produced by the SCAR/piwi cis-NAT in OSS cells. The complementary regions of the SCAR/piwi 3' UTRs specifically generate siRNAs; however, piRNAs are produced from along the length of the SCAR and piwi 3' UTRs. piwi coding exons also generate piRNAs, but fewer than does its 3' UTR.
Although S2 and OSS cells are divergent in their overall gene expression profile, many cis-NAT-siRNA loci were common to OSS cells and S2 cells [20–22]. In fact, 17 of the 40 most highly expressed cis-NAT-siRNA loci were shared by these cell types (Supplementary Table 1B), including CG5148/cher (4th highest-expressed in OSS, #8 in S2), MED21/CG40351 (#5 in OSS, #1 in S2), cenB1A/CG31365 (#6 in OSS, #14 in S2), and CG7739/AGO2 (#7 in OSS, #3 in S2). These data are consistent with the proposal that there exist preferred cis-NAT substrates for the endo-RNAi pathway [20]. Curiously, the third most highly-expressed cis-NAT-siRNA locus in OSS cells was piwi/SCAR (Figure 1C); thus, transcripts for two Argonaute effectors are leading substrates of endo-RNAi in Drosophila.
While the reads from S2 cis-NAT overlaps were strictly 21 nt in length, OSS cis-NAT overlap reads exhibited an unexpectedly bimodal size distribution reflecting both siRNAs and piRNAs (Figure 1B). As with other cis-NATs, the SCAR/piwi locus illustrated that piRNA production was not confined to the region of double-stranded transcription, as is characteristic of endo-siRNA production. Instead, piRNAs preferentially mapped to the sense strands of both SCAR and piwi 3' UTRs, and to a lesser extent from the piwi coding region. Such patterns suggested that primary piRNAs derive from independent consumption of transcripts produced from either strand. The spatially segregated registers of piRNA and siRNA production within 3' UTRs indicated that, in addition to being translated, subpopulations of a transcript could enter multiple small RNA biogenesis pathways.
On the basis of these observations, we conducted a transcriptome-wide survey for piRNAs derived from annotated protein-coding transcripts, yielding 1.25 million such piRNAs out of 13.8 million total library reads (Figure 1C). Thus, the mRNA-directed primary piRNA pathway is highly active: the production of 3' UTR-piRNAs is ~22% the amount of TE-piRNAs and ~30% the amount of miRNAs in OSS cells [11]; mRNA-derived piRNAs outnumbered cis-NAT-siRNAs by 40-fold. This pathway is also broadly active: at an expression cutoff of >50 piRNAs, we defined 2356 mRNAs that generated 1.1 million piRNAs (Supplementary Table S2). This collection comprised >12% of Drosophila transcripts, compared to ~2% of transcripts that generated cis-NAT-siRNAs.
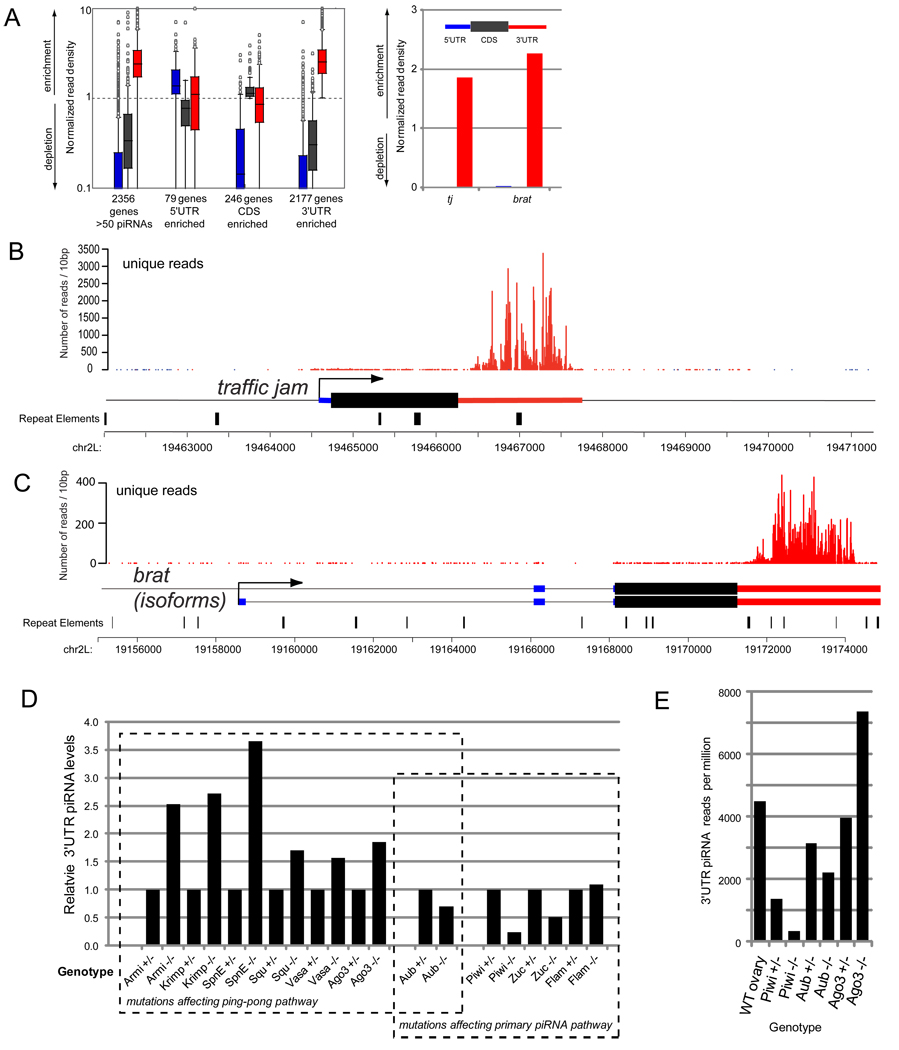
Over 70% (877,808) of mRNA-derived piRNAs mapped to 3' UTRs (Figure 1C), even though the aggregate length of 3' UTRs was much smaller than coding regions (5.4Mb vs. 22.5Mb). The strong bias for 3' UTR-directed piRNA production was illustrated by plotting the cumulative relative density of piRNAs derived from 5' UTRs, CDS and 3' UTRs (Figure 2A). Despite this strong bias, a small subset of mRNAs preferentially generated piRNAs across their coding regions (Figure 2A).
Figure 2.
Preferred production of piRNAs from 3' UTRs. (A) Normalized piRNA read density amongst the transcript features of 5’ UTRs, CDS and 3’ UTRs in genes with >50 piRNAs from OSS cells. Values greater than 1 indicate enrichment of piRNAs, values less than 1 indicate a depletion (see Methods). The box plot on the left shows the distribution of read density values amongst the group of genes with >50 piRNAs, and a breakdown of genes enriched in certain features. The 3’ UTR feature contains the majority of genes and the greatest overall enrichment values. The bar graph on the right shows the enrichment of piRNAs in the 3’ UTR of tj and brat. 5’ UTRs, CDS and 3’ UTRs features are represented by blue, dark grey, and red colors, respectively. (B,C) Example of 3' UTR-directed piRNA production from the traffic jam and brat transcripts. (D) Comparison of proportions of 3’ UTR piRNAs in small RNA libraries from heterozygous and homozygous Drosophila mutants. 3’ UTR levels in homozygotes were normalized to the levels in the corresponding heterozygotes. (E) Normalized 3' UTR piRNA reads per million in wild type and Piwi-class mutants. piwi heterozygous ovaries exhibit substantially lower 3' UTR piRNAs than to aub or AGO3 heterozygotes.
The upper levels of piRNA accumulation were considerable: 180 genes generated >1000 piRNAs (Supplementary Table S2 and Supplementary Figure S1). The loci with the most abundant 3' UTR piRNAs were traffic jam (tj) and brain tumor (brat) with >67,000 and >22,000 piRNAs, respectively, and their piRNAs covered the entirety of these 3' UTRs (Figure 2B,C). tj encodes a Maf-bZIP transcription factor that is specifically expressed in somatic gonadal cells and genetically required for ovary and testis development [24]. brat is a translational regulator required for development and growth control [25, 26], and has also been implicated in the miRNA pathway [27]. The 3' UTR-directed piRNA pathway produced levels of small RNAs from some transcripts that were equivalent to reasonably highly-expressed miRNAs (e.g., only 13 miRNA genes generated more reads than tj, Supplementary Table S2).
Drosophila 3' UTR piRNAs are produced by a primary pathway dependent on Piwi
To gain evidence for exonic piRNA production in the animal, we analyzed large-scale data of small RNAs from total ovaries and 0–2 hour embryos published by Hannon and colleagues [14]. We tallied ~100,000 such piRNAs in the combined available ovary data, and ~65,000 piRNAs in the combined embryo data. When normalized for the total reads in each dataset, OSS cells expressed considerably higher levels of 3' UTR-derived piRNAs, ~56,800 reads per million (RPM) vs. 4,500 RPM in ovaries and 3,000 RPM in early embryos. We defined 316 and 253 transcripts in the ovary and embryo, respectively, which crossed a normalized threshold equivalent to the 50 piRNA cutoff for OSS cells. Importantly, the strong majority of 3' UTR piRNA-generating transcripts detected in the animal were shared by OSS cells (Supplementary Figure S2A), indicating that this reflects a normal pathway rather than an aberrant feature of cultured OSS cells. The 12.6-fold elevation of 3' UTR-derived piRNAs in OSS cells was in line with our observation that follicle-cell specific flamenco piRNAs are ~8-fold higher in OSS than in total ovaries [11], suggesting that 3' UTR-piRNAs are preferentially generated in follicle cells. Similar to primary TE-piRNAs, 3' UTR-derived piRNAs exhibited a 5' U bias (66% of 3' UTR-piRNAs), which is also consistent with their production by a primary piRNA pathway.
The Hannon and Zamore groups recently sequenced ovarian small RNAs from heterozygous or homozygous mutants in a variety of piRNA pathway components [28, 29]. Hannon and colleagues observed that all piRNA pathway mutants exhibited decreased levels of TE-piRNAs, but that only piwi, zucchini and flamenco specifically decreased levels of flamenco piRNAs [28]. These and other observations supported a model in which germline cells (but not follicle cells) engage in strong ping-pong amplification, and that zucchini and piwi are uniquely involved in a primary piRNA biogenesis pathway in somatic ovarian cells.
We collected the mRNA-derived reads from these datasets and observed that 3' UTR-derived piRNAs (and coding exonic piRNAs to a lesser degree) exhibited many similarities to TE-piRNAs across the mutants. For example, zucchini and piwi mutants were strongly decreased for mRNA-derived piRNAs (Figure 2D), consistent with their role in primary TE-piRNA biogenesis. flamenco did not substantially alter 3' UTR piRNA population, befitting its status as a piRNA substrate rather than a biogenesis component per se. In contrast, the ping-pong factors armi, krimper, spn-E, squash and vasa exhibited increased proportions of 3' UTR piRNAs. However, this does not necessarily reflect that these genes limit the accumulation of 3' UTR piRNA. We also observed that the mutants with the strongest reduction in piRNAs from the ping-pong cluster 42AB--armi, spn-E and krimper [28]--correspondingly exhibited the highest proportions of 3' UTR-piRNAs (Figure 2D). We hypothesize that the absence of TE-piRNA amplification may incidentally lead to higher representation of 3' UTR piRNAs in the remaining small RNA pool.
Examination of the other Piwi-class mutants, aub and AGO3, was also informative [28, 29]. While AGO3 mutants had increased proportions of 3' UTR piRNAs, similar to most other ping-pong mutants, aub actually exhibited decreased 3' UTR piRNAs. This suggested that Aub received some 3' UTR piRNAs via a primary biogenesis pathway operative in the germline, consistent with the notion that Aub and Piwi are both loaded with primary piRNAs while AGO3 mostly contains secondary piRNAs [2, 3]. Still, the piwi mutation clearly had greatest effect on 3' UTR piRNA accumulation. Indeed, piwi exhibited substantial haploinsufficiency, since piwi heterozygotes accumulated far fewer 3' UTR piRNAs than did aub or AGO3 heterozygotes (Figure 2E). In summary, these analyses support the existence of a primary piRNA biogenesis pathway in vivo, with specificity towards the 3' UTRs of protein-coding transcripts and marked dependence on PIWI.
Conservation of 3' UTR piRNAs in vertebrates
We next assessed whether the mRNA/3' UTR-directed piRNA pathway was apparent in mammalian gonads. We analyzed published mouse pre-pachytene testes (10 dpp) data [4, 30] and generated new small RNA data from post-pachytene (8 weeks adult) testes. The proportion of pre-pachytene spermatocytes in relation to other testicular cell types is highest at 10 dpp; subsequently, post-pachytene spermatocytes and spermatids dominate in the testes [34, 35]. At 10 dpp, Mili appears to be sole Piwi protein expressed, whereas the adult testis expresses both Mili and Miwi [4].
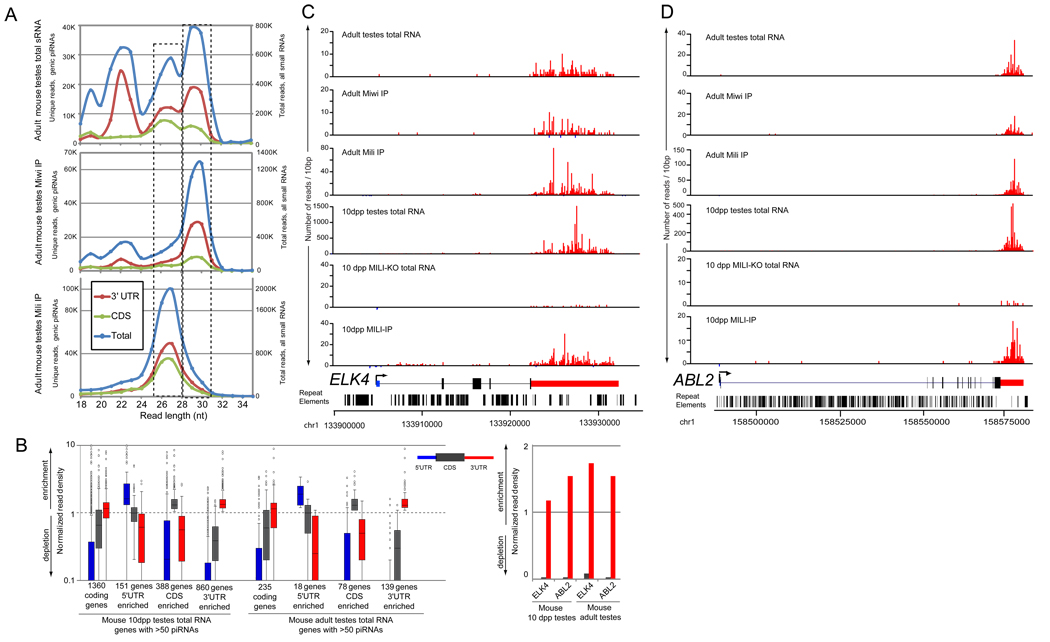
Exonic piRNAs have been noted [4, 8, 10, 30, 33], but their regional preferences within protein-coding transcripts were not previously characterized in detail. Examination of previously published lists of the top 200 fetal, 100 pre-pachytene, and 94 post-pachytene clusters [4, 10, 36], indicated that many of these clusters could be re-classified as 3’ UTR-directed piRNA clusters (Supplementary Table S3; e.g. clusters ranked #34 and #2 in fetal and 10dpp testis, respectively, are piRNAs against the 3’ UTR of ELK4, while adult testis cluster ranked #49 has piRNAs against the 3’ UTR of CBL). As was the case in Drosophila follicle cells, we observed substantial bias for 3' UTR-directed piRNA production in mouse testes (Figure 3B), although there were distinct populations of transcripts with apparent CDS or even 5' UTR enrichment for piRNA production. 3' UTR-piRNAs were seemingly much more abundant at 10dpp (~35% of all library reads) than in the adult (1.8% of all reads), although their representation in the adult library may incidentally be lowered due to the tremendous output of pachytene piRNA clusters [7–10].
Figure 3.
Mammalian 3' UTR-directed piRNAs and representative directed transcripts. (A) Size distribution of reads sequenced from total small RNAs (top graph), a MIWI IP (middle graph), and a MILI IP (bottom graph) from adult mouse testes extract. Dotted boxes highlight congruent peaks between the graphs. (B) Normalized piRNA read density amongst the transcript features of 5' UTR, CDS, and 3' UTR in coding genes with >50 piRNAs from mouse 10dpp and adult testes. Values greater than 1 indicate enrichment of piRNAs; values less than 1 indicate a depletion (see Methods); and 5’ UTR, CDS and 3’ UTR features are represented by blue, dark grey, and red colors, respectively. The box plot on the left shows the distribution of read density values amongst the group of coding genes with >50 piRNAs, and a breakdown of genes enriched in certain features. In line with OSS cell genic piRNAs, the 3’ UTR feature contains the majority of genes and the greatest overall enrichment values. The bar graph on the right shows the enrichment of piRNAs in the 3’ UTR of ELK4 and ABL2. Hundreds of murine transcripts, such as ELK4 (C) and ABL2 (D), preferentially generate piRNAs from the sense strand of their 3' UTRs (see also Supplementary Figure S3). Repeats track indicates that 3' UTRs are strongly depleted in transposon/repetitive sequences.
From combined data in 10dpp and adult testes total RNA libraries, we identified 829 transcripts 50 or more sense-oriented, uniquely mapping piRNAs, with 83 transcripts generating >1000 piRNAs (Supplementary Figure S3 and Supplementary Table S5A and S5B). For abundant piRNA-generating mRNAs, the strong majority of piRNAs were located in 3' UTRs (424,227 uniquely mapping reads) relative to 5' UTR and CDS (55,799 total uniquely mapping reads). Even following normalization for gene lengths, piRNA density was highest amongst 3' UTRs in both 10dpp and adult testes libraries (Figure 3B). The Ets-domain oncogene family gene, ELK4, and the Abelson murine leukemia viral oncogene homolog 2, ABL2, serve as typical examples of transcripts with 3' UTR-directed piRNA production (Figure 3C–D). The enrichment of 3' UTR piRNAs is actually more considerable since ~1/3 of the reads mapping outside of 3' UTRs derived from non-coding RNAs like Rnu1b2, Snord22 and Gas5, whose abundant reads came from point-sources (Supplementary Figure S4); or genes like Fth1, which are transcripts with abundant piRNA production from 5' UTR and CDS (Supplementary Figure S5).
Analysis of a published 10 dpp library from mili −/− testes [4] revealed a strong depletion of genic piRNAs, including at 3' UTRs (Figure 3 and Supplementary Figure S3). This genetic dependence echoed the observation that 3' UTR-piRNAs are highly dependent on Drosophila piwi. We observed a strong 5' U bias (>85%) for murine mRNA/3' UTR-derived piRNAs, suggesting that these are primary piRNAs as in flies.
Loading of 3' UTR-derived piRNAs into Piwi complexes in three animal clades
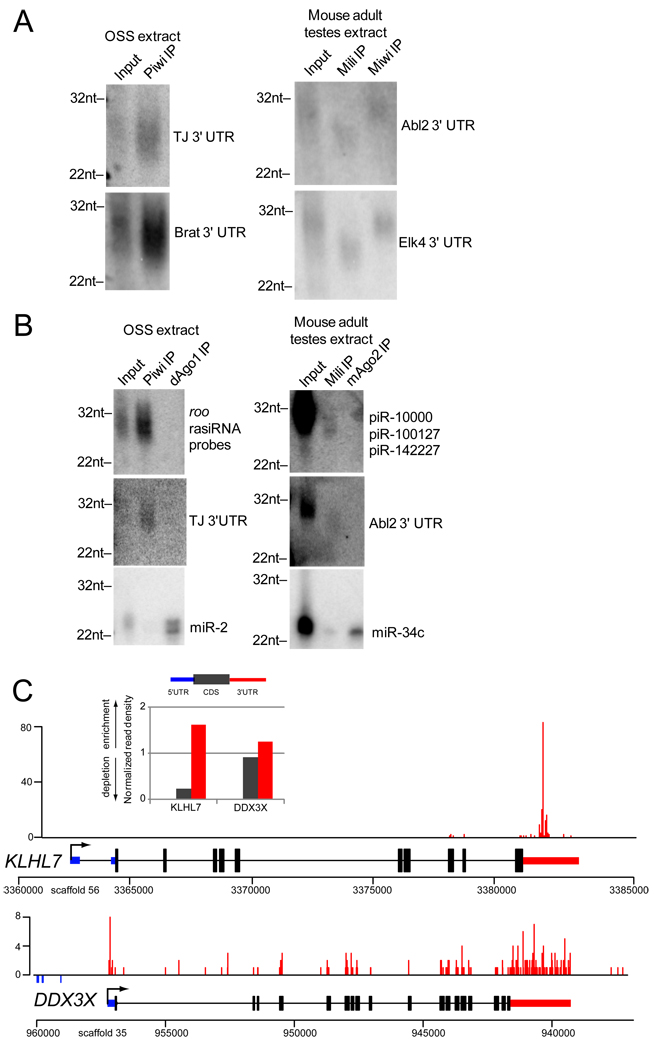
We next sought biochemical validation of the loading of 3' UTR piRNAs into Piwi complexes. Analysis of Piwi immunoprecipitates (IP) from OSS cells revealed that it carried piRNAs derived from the tj and brat 3' UTRs, but did not contain the microRNA miR-2 (Figure 4). Reciprocally, AGO1-IP contained miR-2 but not 3' UTR piRNAs. In adult mouse testes, we detected Abl2 and Elk4 piRNAs in both Mili and Miwi complexes, but not in mAGO2 complexes. Equivalently-sized probes directed at coding exons of these genes did not yield signal (data not shown), consistent with the much lower abundance of coding piRNAs from these mRNAs.
Figure 4.
Association of 3’ UTR-directed piRNAs in Piwi-protein complexes. (A) RNAs from PIWI, MILI, and MIWI IPs were probed on Northern blots with cloned 3’ UTR fragments. (B) The 3’ UTR probes are specific for piRNAs from Piwi-protein IPs. (C) Deep conservation of 3’ UTR-directed piRNAs in animals. Enrichment of piRNA reads to the 3’ UTR of KLHL7 and DDX3X genes from an IP of Xenopus Piwi proteins with the Y12 antibody, as illustrated by the gene map and the bar graph of normalized piRNA read density according to transcript feature. 5’ UTR, CDS and 3’ UTR features are represented by blue, dark grey, and red colors, respectively.
Curiously, the peak size of 3' UTR piRNAs in Mili complexes (27–28 nt) was distinctly shorter than those in Miwi complexes (29–30 nt), reminiscent of the observation that different Piwi proteins are associated with characteristic sizes of piRNAs [3, 30, 32]. The sizes of the Northern blot signals from the Miwi and Mili IP were in concordance with deep-sequencing analysis (Figure 3A). In addition, the observation that bulk 3' UTR piRNAs in total RNA were the same size as those in Miwi-IP suggested that Miwi is the predominant carrier of 3' UTR piRNAs in adult testes. On the other hand, Mili may be the primary carrier of 3’UTR piRNAs in 10dpp testes because Miwi is not yet expressed at this stage [4, 30]. Consistent with this, a previously reported 10 dpp Mili-IP library contained 3' UTR-biased distribution of 26–27 nt piRNAs, similar to total RNA data from this stage [4].
We confirmed the distribution of 3' UTR-derived piRNAs by sequencing 8.2 million Mili-IP and 5.5 million Miwi-IP mapped reads from adult testes, and comparing these to our matched total RNA reads. To our knowledge, Mili-IP has not previously been deeply sequenced at this stage, and no deep Miwi-IP data have been reported thus far. We observed that the total RNA library contains three distinct size peaks at 22 nt, 27 nt and 30 nt (Figure 3A). The former corresponds to miRNA reads, while the latter two peaks correspond to piRNA reads. Consistent with the Northern blots, analysis of the IP libraries showed that these peaks segregated precisely with the contents of Mili and Miwi complexes, respectively. In addition, the total RNA library exhibited greater representation of 3' UTR reads whose sizes corresponded to Miwi complexes, as we inferred from Northern analysis. When focusing on reads that matched to 3' UTRs, we observed similar trends across these libraries, indicating that 3' UTR piRNAs were loaded into distinct Piwi-class complexes in adult testes. Nevertheless, the genes hit by piRNAs were relatively similar between Mili- and Miwi-IPs (data not shown), suggesting their production via a common primary pathway.
To address whether 3' UTR piRNAs might be present in Piwi complexes of mature germ cells in an evolutionarily distant vertebrate outgroup species, we examined in Xenopus tropicalis eggs piRNAs thatassociated with Y12 antibody, which binds symmetrically methylated arginines [37] that are present on diverse Piwi proteins [32, 38]. Despite incomplete genome annotation and the high proportion of reads from intergenic regions and TEs, we identified numerous annotated mRNAs with piRNAs corresponding to exons and 3' UTRs (Figure 4C, data not shown). These reads were less abundant than in mouse and Drosophila, but KLHL7 and DDX3X serve as examples for piRNA enrichment at 3' UTRs. KLHL7 is implicated in ubiquitination and is associated with retinitis pigmentosa [39], while DDX3X is a helicase homologous to Drosophila Belle, which has been implicated as a component in the endo-RNAi pathway [40]. The existence of 3' UTR piRNAs in Xenopus provides evidence that this biogenesis pathway has been quite broadly conserved amongst animals.
Selective production of 3' UTR-directed piRNAs from gonadal mRNAs
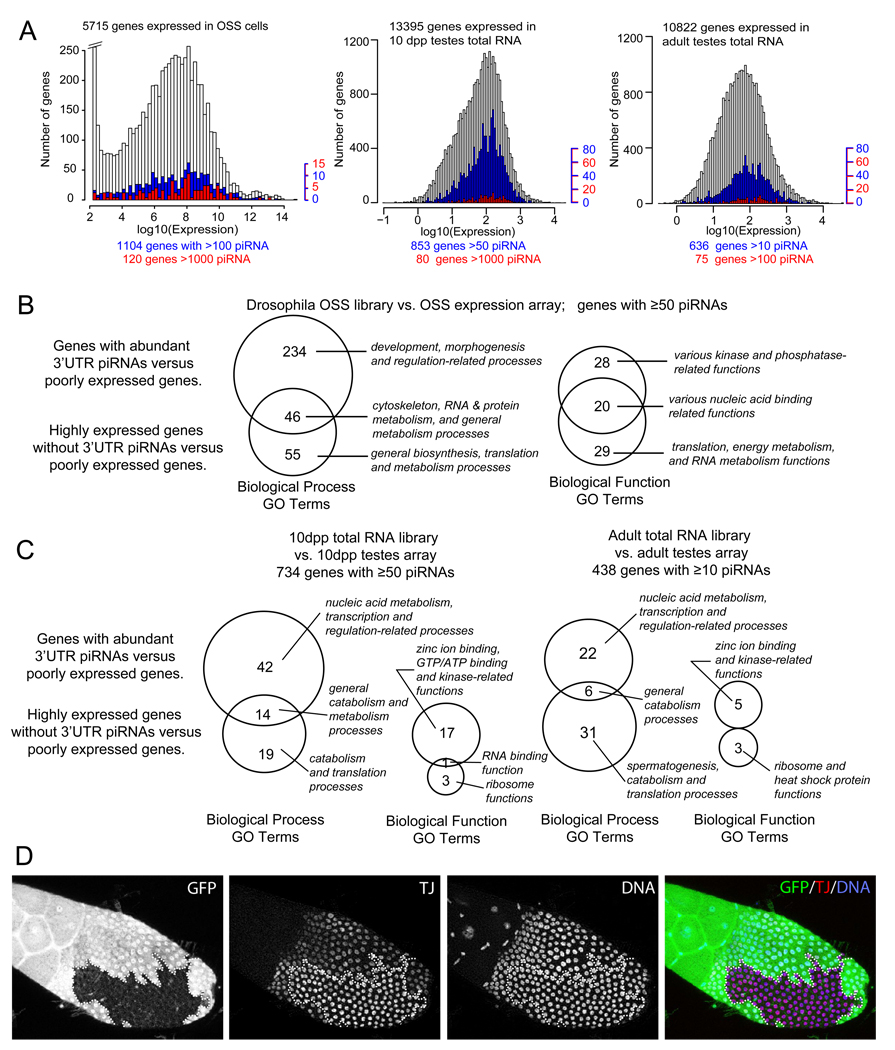
We next assayed the relationship of piRNA-producing transcripts with Affymetrix gene expression data from OSS cells (this study) and murine testis [41]. Comparing expression of mRNA generating 3’ UTR-piRNA with the general population, we observed that transcripts with moderate to high level of piRNA were biased for higher expression (mean log2 expression=6.7 for all transcripts called present, 7.3 for mRNAs with >100 piRNAs and 7.4 for mRNAs with >1000 piRNAs, Supplementary Figure S2B). Nevertheless, a large population of highly expressed transcripts (385 transcripts with log2 expression value of >8 did not generate piRNAs, indicating that piRNA production was not strictly determined by transcript abundance. This was illustrated by plotting the gene expression of all mRNAs vs. those that generated >100 piRNAs and >1000 piRNAs (Figure 5A). One might have expected transcripts with higher piRNA production to be greatly skewed towards higher expression levels; instead, the mRNAs with highest piRNA production spanned the full range of transcript accumulation.
Figure 5.
(A) Histograms of genes by their ranked log10 expression levels and their corresponding count of 3’ UTR piRNAs from OSS cells, mouse 10dpp testes total RNA, and mouse adult testes total RNA libraries. Expressed genes are represented in white bars, genes with noted cutoff of piRNAs are in blue or red bars. (B) Venn diagrams showing the comparison of GO term enrichments in mRNAs with abundant 3' UTR-directed piRNAs from OSS cells. Shown are counts of GO terms enriched amongst mRNAs with abundant (>1000 reads) and preferred production of piRNAs from 3' UTRs. Call-out labels highlight notable GO terms with the strong statistical significance (p<10−4). (C) Venn diagrams comparing GO term enrichments between mRNAs with abundant 3’ UTR-directed piRNAs and highly expressed mRNAs lacking 3’ UTR piRNAs in mouse 10dpp and adult testes. Details of the GO analyses, including of piRNA-producing transcripts lacking preferred 3' UTR enrichment, are available in Supplementary Tables S6A–B (fly) and S7 (mouse). (D) Staining of piwi mutant follicle clones with GFP antibody (green, to identify clone borders), Tj antibody (red) and DAPI (blue, to identify nuclei). Homozygous piwi mutant clones, as marked by absence of GFP, exhibit elevated levels of Tj.
We found the same to be true for 10 dpp and adult mouse testes. Although there was a slight skew for piRNA-producing transcripts towards higher steady-state levels, they exhibited a similar distribution as the profile of all genes that are expressed in mouse testes (Figure 5A). These data suggest that the primary piRNA pathway selects the 3' UTRs of transcripts across a broad expression range, rather than acting non-specifically on transcripts in proportion to their abundance.
Selection of transcripts with specific GO enrichments by the primary piRNA pathway
Since there appeared to be selectivity for mRNA substrates by the primary piRNA pathway, we tested for enrichment of specific gene ontology (GO) categories. We first analyzed 646 OSS mRNAs that generated >200 3' UTR piRNAs and had GO terms (regardless of gene expression level), relative to a background set of ~3000 lowly-expressed OSS transcripts with GO terms. To assess whether piRNA-producing genes exhibited properties that were distinct from merely abundant transcripts, we analyzed the 675 most abundant OSS mRNAs that produced at most 10 piRNAs. These gene sets overlapped modestly in their GO terms, which included cytoskeleton, RNA and protein metabolism processes and various nucleic acid binding-related functions (Figure 5B). However, the themes amongst GO terms exclusively represented in either dataset were notably different: abundant genes lacking piRNAs were most highly enriched in "housekeeping" GO categories such as general biosynthesis, metabolism, translation and ribosome components, and general transcription machinery. In contrast, the most highly enriched GO terms amongst genes with abundant 3’UTR piRNAs included categories such as development, morphogenesis and regulatory processes, as well as DNA binding proteins, kinases and phosphatases. The specific enrichment of many regulatory and developmental functions amongst abundant piRNA-generating genes, but not amongst abundantly expressed genes per se, suggests possible regulatory coherence by the primary piRNA pathway.
To analyze the murine data, we used cutoffs of ≥50 piRNAs/mRNA in 10 dpp total RNA library, and ≥10 piRNAs/mRNA in total RNA, Mili-IP and Miwi-IP in adult testis libraries (to account for lower 3’ UTR piRNA content in adult libraries). Equivalently-sized cohorts of mRNAs with high 3' UTR piRNAs and a non-overlapping set of the highest expressed mRNAs without piRNAs revealed no overlap in enriched GO Function categories and only a modest overlap in Process categories (Figure 5C), indicating that genes that generated abundant piRNAs were functionally distinct from abundant transcripts. Terms highly enriched in abundant transcripts without piRNAs at both 10dpp and adult testes included many housekeeping categories, such as catabolism, translation and ribosome components. On the other hand, terms that were highly and uniquely enriched amongst abundant piRNA transcripts included transcription and nucleic acid metabolic processes, and zinc ion-binding and kinase-related functions.
Taken together, these analyses clearly demonstrate that abundant piRNA-producing transcripts in Drosophila OSS cell and murine testis comprise gene cohorts whose functions are very distinct from those of highly-expressed transcripts in either species. Interestingly, several of the GO categories enriched in piRNA-producing transcripts were shared between fly and mouse gonads, even though the actual genes involved were quite different. We take this as a strong suggestion that the selection of mRNAs encoding certain types of GO functions is important to the operation of the 3' UTR-directed piRNA pathway. Interestingly, posttranscriptional gene silencing by RNA was a category enriched amongst abundant piRNA-generating mRNAs in both OSS cells and adult murine testes, but not amongst abundant transcripts in either tissue. In Drosophila, these genes included brat, Dcr-2, RM62, AGO1 gawky/GW182 and piwi (Supplementary Table S2), while in mouse these included TNRC6b, eif2c2/mAgo2, eif2c4/mAgo4, and piwil2/mili (Supplementary Table S5A, S5B).
Beyond the small subset of small RNAs that coincidentally derive from cis-NATs or are related to TEs (e.g. Figure 1D and Figure 3), the bulk of mRNA/3' UTR-derived piRNAs lack highly complementary sequences in the transcriptome. Although some mRNA/3' UTR-derived piRNAs conceivably guide the regulation of trans-encoded targets bearing imperfect matches, a more proximal effect is the removal of 3' UTRs from transcripts via abundant piRNA production. However, we did not observe substantial, consistent changes in the average expression of murine piRNA-producing mRNAs in mili +/− vs. −/− testes (Supplementary Table S8).
In principle, the results from whole gonads might be confounded if only a subset of cells have the capacity for piRNA generation, and/or if there is a discrepancy between mRNA and protein output. We examined this in more detail by generating piwi mutant clones in Drosophila ovaries and staining them for TJ, whose mRNA generated the most abundant 3' UTR piRNAs in OSS cells. We observed many clones in which TJ protein was upregulated in piwi mutant cells relative to neighboring control cells (Figure 5D). There appeared to be a temporal dependence in that younger clones did not exhibit increased levels of TJ, perhaps because of insufficient time for its deregulation. Nevertheless, these data support the notion that this pathway might influence target output.
Discussion
Genesis of abundant primary piRNAs from 3' UTRs in diverse animals
Our observations highlight the prevalence and conservation of mRNA substrates for the primary piRNA pathway, and provide insight into primary piRNA biogenesis. Although clear orthologs between mouse and Drosophila Piwi proteins are not established, germline-associated cells in both systems engage a primary piRNA biogenesis pathway that preferentially generates abundant piRNAs from 3' UTRs. This class of piRNAs is especially abundant in Drosophila follicle cells (via Piwi), and in pre-pachytene (via Mili) and pachytene (via both Miwi and Mili) stage mammalian testes; we also detected a similar Aub-dependent pathway that likely operates in the Drosophila germline. While this work was under review, Siomi and colleagues similarly reported the existence of a piwi- and zucchini-dependent primary piRNA pathway that operates on the 3' UTRs of some Drosophila mRNAs [42]. Our conclusions on biogenesis are generally consistent. However, as our sequence data were ~1000 fold deeper, we could demonstrate that the primary piRNA pathway is not selective for a few mRNAs, but instead acts broadly across >1000 cellular transcripts, and operates in both flies and vertebrates.
The mechanism of primary piRNA biogenesis remains largely mysterious. We believe that mRNA/3' UTR-directed piRNA generation results from a primary processing pathway, based on their 5' U bias, their extraordinary abundance in Drosophila OSS cells (which lack ping-pong), and their genetic independence from ping-pong components. It is worth mentioning that previous studies noted that some 3' UTRs harbored TE sequences, suggesting a potential relationship to TE-piRNA production [4, 30]. However, we observed no overall enrichment for TEs in the 3' UTRs of piRNA-generating mRNAs (data not shown). Indeed, many Drosophila and mammalian 3' UTRs with highest piRNA production were strongly depleted for repeat elements relative to their introns (e.g. Figure 2 and Figure 3), and most mRNA-derived piRNAs mapped uniquely. Moreover, the clearly distinct patterns of siRNA and piRNA biogenesis within cis-NAT-3' UTRs, along with the general observation that most abundant piRNA-generating transcripts are not arranged in cis-NATs, suggest that the endo-siRNA and primary piRNA pathways act independently on cellular transcripts.
The production of piRNAs from spliced transcripts and preferentially from 3' UTRs suggests that primary piRNA processing may be a cytoplasmic event occurring on transcripts engaged with ribosomes. This scenario is consistent with the co-sedimentation of Miwi and Mili with polysome fractions [9, 43]. Our data do not rule out the existence of 3' UTR-specific transcripts that generate piRNAs, but we did not detect accumulation of stable 3' UTR-specific transcripts from loci with highly abundant 3' UTR piRNAs (data not shown). Instead, we speculate that the abrupt reduction of piRNA production from adjacent coding exons might reflect competition between the translation machinery and primary piRNA biogenesis machinery. We analyzed a set of putative non-coding loci and while their status as non-coding has not generally been examined experimentally, many of them exhibited distributive patterns across their exons (Supplemental Figure S4). On the other hand, we note that a subset of protein-coding genes exhibit substantial CDS-piRNAs (e.g. piwi, Figure 1). A careful comparison of mRNA partitioning between free and translating pools with their distribution of piRNA reads would be an informative test of the competition model.
The Tudor-domain containing gene TDRD-1 interacts with Miwi and Mili [32], and was suggested to regulate the formation of intergenic piRNAs versus genic piRNAs [33]. Our analysis confirmed that genic piRNAs derived predominantly from 3' UTR piRNAs were elevated in the 15 dpp TDRD-1 KO library comparable to 10dpp WT libraries (Supplemental Table S4B). However, we did not observe a similarly high proportion of 3' UTR piRNAs in a TDRD-1 null 18.5 dpc testis [32]. We note that TDRD-1 mutants exhibit apoptotic post-pachytene spermatocytes at 15 dpp [44], and suggest that this reduces the contribution of post-pachytene intergenic piRNAs in total small RNA libraries. This should concomitantly increase the proportion of 3' UTR piRNAs, thereby providing an alternate explanation for their increase besides a direct role for TDRD-1 in piRNA substrate selection [33].
The existence of abundant 3' UTR-derived piRNAs raises the question of a potential relationship with pyknons. These are n-mers (exhibiting characteristic lengths ranging from 18–31 nt) that are over-represented in intergenic space and have additional matches in genic space [45]. Curiously, genic instances of pyknons are concentrated in 3' UTRs, and at least some of the GO terms that are enriched amongst transcripts with pyknon instances are shared by transcripts that generate abundant 3' UTR piRNAs (such as transcription and nucleic acid metabolism). However, 3' UTRs that are metabolized by the primary piRNA pathway broadly produce piRNAs from across their lengths with relatively little specificity for individual piRNAs, and the vast majority of these piRNAs do not match intergenic sequences (beyond the small fraction that contain repetitive sequences of TEs). These properties suggest that the primary piRNA pathway metabolizes 3' UTRs independently of pyknon instances, although potential functional intersection between these phenomena requires additional study.
Biological function of the 3' UTR-directed primary piRNA pathway
A challenge for the future is to understand the biological consequences of the 3' UTR-directed primary piRNA pathway. Since piRNA-generating transcripts were not consistently altered in mili-KO mutants, it is prudent to consider whether these piRNAs might be incidental. We do not rule this out, and perhaps some are byproducts of poised TE defense operation. On the other hand, it seems difficult to imagine that they entirely lack regulatory consequences given their sheer abundance: 3' UTR piRNAs comprise nearly 10% of small RNAs in Drosophila OSS cells and 35% of small RNAs in 10 dpp mouse testis.
Given the breadth of mRNA substrates, it is also conceivable that the primary piRNA pathway selects its substrates indiscriminately, perhaps in an attempt to identify selfish genetic material. However, this scenario is not reconciled with the observation that very substantial populations of abundant transcripts in Drosophila and mouse gonads do not generate 3' UTR piRNAs. Therefore, the piRNA pathway can no longer be seen simply as having to choose between self (endogenous transcripts) and non-self (i.e. TEs). Instead, it must further route specific populations of host transcripts for 3' UTR-directed piRNA production, which we hypothesize has regulatory consequences.
Such regulation may operate in cis or in trans. In support of the former scenario, the tj 3' UTR is one of the most abundant sources of mRNA-derived piRNAs, and we observed increased levels of the transcription factor TJ in piwi mutant clones (Figure 5D). Perhaps more compelling was the finding that mRNAs selected for abundant 3' UTR piRNA production were enriched in a variety of GO categories that were not representative of abundant transcripts per se. We take this as an indication that they were actively selected for piRNA production. Moreover, the fact that many GO categories were shared by abundant piRNA-generating transcripts between Drosophila and mouse, such as kinase-related factors, DNA binding factors, and RNA silencing factors, suggests that they represent functionally conserved targets of the primary piRNA pathway. While experimental evidence for alteration of small RNA pathway components via piRNA generation remains to be demonstrated, these putative auto-regulatory connections may expand a theme that includes the targeting of mammalian Dicer by let-7 [46, 47], the cleavage of DGCR8 by Drosha [48], the existence of abundant endo-siRNAs emanating from Drosophila AGO2 [20–22], the regulation of plant AGO1 and Dicer-like 1 by various miRNAs [49–51], and the regulation of a dsRNA intermediate that generates the RNAi regulator ERI-6/7 [52].
It is also conceivable that mRNA/3' UTR-derived piRNAs serve as guides to regulate the expression of other transcripts. While this work was under review, Siomi and colleagues proposed that tj-derived 3' UTR piRNAs regulate cell adhesion genes such as fas III, potentially via partially complementary sequences located in its ~60kb intron [42]. Our data do not directly address this model; however, the candidate fas III-targeting tj piRNAs account for only 2.7% of tj piRNAs and only 0.15% of all 3' UTR-piRNAs in our OSS data. Genetic tests involving modification of the endogenous tj 3' UTR may clarify whether these rare specific tj piRNAs indeed serve critical trans-regulatory roles. Nonetheless, in light of the great abundance of mRNA/3' UTR-piRNA complexes in Drosophila and mouse gonads, we consider it plausible that they collectively have substantial trans-regulatory impact. In any case, the revelation of broad and abundant generation of piRNAs from cellular transcripts raises a new direction to understand how the piRNA pathway influences gonad and germ cell development.
Materials and Methods
Generation and analysis of small RNA data
Testes extract from 8 week old Swiss Webster mice (Pel-Freez Biologicals) was prepared as described [10]. Miwi and Mili IPs were performed using 20ug of antibody bound to 100 µl of Protein A Dynabeads (Invitrogen) for 2 hrs at 4°C, followed by 4 washes with 1X PBS and 0.5% NP40, 0.5% Triton-X100. Y12-IP from Xenopus tropicalis egg extract was performed as described [53]. Small RNA libraries from total RNA or IP material were constructed and subjected to Illumina sequencing [11]. Small RNA library data from adult mouse testes and Y12-IP from X.tropicalis egg extract are under submission at the NCBI Gene Expression Omnibus (GEO), and will be publicly available upon publication of this study.
Small RNA reads from OSS cells were described in a separate analysis [11], and the data were deposited at NCBI Gene Expression Omnibus (GEO) under series GSE15378. We also used several small RNA datasets from Drosophila ovary and embryo published by the Hannon and Zamore groups [14, 28, 29], as well as data from the Hannon and Pillai groups from mouse testis and ovary [4, 30–33], all obtained from NCBI-GEO or SRA. To provide uniformity in our analysis, we processed the raw NCBI datafiles along with our own small RNA data by clipping and mapping reads relative to gene annotations in D. melanogaster dm3/Release 5.2, and Mus musculus mm9/Build 37. We only used those reads that matched perfectly to the genome, with no internal mismatches permitted.
Similar to our previous analysis of cis-NAT-siRNAs [20], we observed a number of piRNA clusters encompassing regions on the sense strand downstream of annotated 3' UTRs. In some cases, it was evident that no 3' UTR was annotated, while in other cases the extension might represent alternative polyadenylation usage. To capture these, we extended 3' UTRs by 100 nt intervals to check for downstream piRNA mappings of similar density to the annotated 3' UTR, trimming the extension so as to include unannotated 3' UTR piRNAs that were continuous with the sense strand of the transcript but did not overlap neighboring genes.
To measure the enrichment of reads in category features of transcripts (5' UTR, CDS, 3' UTR), we derived a normalized metric of (# piRNA reads in the category / total # piRNA reads mapping to gene) / (# bases in the category / total # bases in the gene), which accounts for longer length categories having more numerous reads. A value of 1 suggests no enrichment of reads in a category, more than 1 indicates relative enrichment, and less than 1 indicates a relative depletion of reads in a category. Genes with CDS lengths >100 bp were considered coding genes, and the remainder were provisionally considered non-coding.
Gene expression and gene ontology analysis
Drosophila gene expression profiling was performed on three independently cultured samples of OSS cells that were grown as previously described [12]. Total RNA was extracted and used for single-amplification target analysis with the Drosophila v.2.0 3' end Gene Expression microarray (Affymetrix). The gene expression data were processed using R software for statistical computing and graphics (http://www.r-project.org/) and GC-RMA normalization. The microarray data were deposited at NCBI-GEO under series GSE15825 and accession numbers GSM397756, GSM397757 and GSM397758. For murine gene expression analysis, we used dataset GSE4742 comparing D10 mili +/− with mili −/− testis [10] as well as data from various testis stages reported by Griswold and colleagues [41].
GO enrichment analysis was performed using GOrilla (http://cbl-gorilla.cs.technion.ac.il/) [54]. The ‘two unranked lists option’ was used to specify the following sets of target and background genes. For each set, the analysis was run once each for the three main GO categories – Biological process, Molecular Function and Cellular Component using the options ‘Process’, ‘Function’ and the ‘Component’ respectively. We compared the 763 (646 with GO terms) genes with ≥200 reads of 3' UTR piRNAs against the 6321 poorly expressed genes in OSS cells, and the 907 (675 with GO terms) higher expressed genes with less than 10 piRNA against this negative set (Supplementary Table 7). We also compared the 438 (425 with GO terms) mouse genes with >10 3' UTR piRNAs against a background set of 2203 poorly transcribed genes, as well as the most strongly expressed 438 (412 with GO terms) mouse genes with no piRNAs against the same background set (Supplementary Table 8).
Northern analysis
A 10cm dish of confluent OSS cells was lysed in the same buffer used for testis extracts. Piwi complexes were immunoprecipitated from this material using 20 µg of Piwi monoclonal antibody affixed to 100µl of Protein G Dynabeads. Mili-IP and Miwi-IP of mouse adult testes extract were performed as described for library construction. 3' UTR and CDS fragments were PCR amplified from reverse-transcribed mRNA from OSS cells or Oregon R genomic DNA, cloned into vector pCR-II (Invitrogen), and transcribed with α-32P-ATP using the Strip-EZ Kit (Ambion). Northern blots were constructed and probed according to [55].
Drosophila clonal analysis
piwi mutant clones were generated in hs-FLP; piwi[2] FRT40A/ubi-GFP FRT40A animals and stained using rat anti-TJ [24], rabbit anti-GFP (Molecular Probes) and DAPI, followed by Cy3-conjugated goat anti-rat and Cy2-conjugated goat anti-rabbit (Molecular Probes).
Supplementary Material
Acknowledgements
We thank Haruhiko Siomi and Mikiko Siomi for the Piwi monoclonal antibody and for discussing results prior to publication. We are also grateful to Dorothea Godt for TJ antibody, to Haifan Lin for piwi stocks, to Kate Abruzzi and Michael Rosbash for reagents and assistance with Affymetrix gene profiling, and Mark Borowsky and the MGH Illumina sequencing core for assistance in library sequencing. This work was partly supported by NIH grant #GM48405 to Robert Kingston and an NIH grant #GM086434 and a Career Award in the Biomedical Sciences from the Burroughs Wellcome fund to M.D.B. Z. J. is a Research Fellow of The Terry Fox Foundation (Award #700132). N.C.L. was funded by a K99 award from the National Institute of Child Health and Human Development and the NIH (K99-HD057298-01). Work in E.C.L.'s group was supported by the Sidney Kimmel Cancer Foundation, the Alfred Bressler Scholars Fund and the NIH (R01-GM083300 and U01-HG004261).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 2.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 3.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 4.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 5.Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RH, Hannon GJ, Draper BW, Ketting RF. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 6.Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006 doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 8.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006 doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 9.Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci U S A. 2006;103:13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA Complex from Rat Testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 11.Lau N, Robine N, Martin R, Chung WJ, Niki Y, Berezikov E, Lai EC. Abundant primary piRNAs, endo-siRNAs and microRNAs in a Drosophila ovary cell line. Genome Res. 2009;19:1776–1785. doi: 10.1101/gr.094896.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niki Y, Yamaguchi T, Mahowald AP. Establishment of stable cell lines of Drosophila germ-line stem cells. Proc Natl Acad Sci U S A. 2006;103:16325–16330. doi: 10.1073/pnas.0607435103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aravin A, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 14.Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung WJ, Okamura K, Martin R, Lai EC. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Current Biology. 2008;18:795–802. doi: 10.1016/j.cub.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 17.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 18.Kennerdell JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the Wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- 19.Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA. Targeted mRNA degradation by double-stranded RNA in vitro. Genes and Development. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okamura K, Balla S, Martin R, Liu N, Lai EC. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila. Nat Struct Mol Biol. 2008;15:581–590. doi: 10.1038/nsmb.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, Zamore PD. Endogenous siRNAs Derived from Transposons and mRNAs in Drosophila Somatic Cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel J, Sachidanandam R, Hannon G, Brennecke J. An endogenous siRNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada T, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 24.Li MA, Alls JD, Avancini RM, Koo K, Godt D. The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila. Nat Cell Biol. 2003;5:994–1000. doi: 10.1038/ncb1058. [DOI] [PubMed] [Google Scholar]
- 25.Sonoda J, Wharton RP. Drosophila Brain Tumor is a translational repressor. Genes Dev. 2001;15:762–773. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank DJ, Edgar BA, Roth MB. The Drosophila melanogaster gene brain tumor negatively regulates cell growth and ribosomal RNA synthesis. Development. 2002;129:399–407. doi: 10.1242/dev.129.2.399. [DOI] [PubMed] [Google Scholar]
- 27.Neumuller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature. 2008 doi: 10.1038/nature07014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, Kittler EL, Zapp ML, Klattenhoff C, Schulz N, Theurkauf WE, Weng Z, Zamore PD. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol. 2009;16:639–646. doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- 34.Bellve AR, Cavicchia JC, Millette CF, O'Brien DA, Bhatnagar YM, Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nebel BR, Amarose AP, Hacket EM. Calendar of gametogenic development in the prepuberal male mouse. Science. 1961;134:832–833. doi: 10.1126/science.134.3482.832. [DOI] [PubMed] [Google Scholar]
- 36.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lerner EA, Lerner MR, Janeway CA, Jr, Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci U S A. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol. 2009;11:652–658. doi: 10.1038/ncb1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedman JS, Ray JW, Waseem N, Johnson K, Brooks MJ, Hugosson T, Breuer D, Branham KE, Krauth DS, Bowne SJ, Sullivan LS, Ponjavic V, Granse L, Khanna R, Trager EH, Gieser LM, Hughbanks-Wheaton D, Cojocaru RI, Ghiasvand NM, Chakarova CF, Abrahamson M, Goring HH, Webster AR, Birch DG, Abecasis GR, Fann Y, Bhattacharya SS, Daiger SP, Heckenlively JR, Andreasson S, Swaroop A. Mutations in a BTB-Kelch protein, KLHL7, cause autosomal-dominant retinitis pigmentosa. Am J Hum Genet. 2009;84:792–800. doi: 10.1016/j.ajhg.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou R, Hotta I, Denli AM, Hong P, Perrimon N, Hannon GJ. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol Cell. 2008;32:592–599. doi: 10.1016/j.molcel.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- 42.Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- 43.Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi-Miyagawa S, Nakano T, Lin H. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J Biol Chem. 2009;284:6507–6519. doi: 10.1074/jbc.M809104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chuma S, Hosokawa M, Kitamura K, Kasai S, Fujioka M, Hiyoshi M, Takamune K, Noce T, Nakatsuji N. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci U S A. 2006;103:15894–15899. doi: 10.1073/pnas.0601878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rigoutsos I, Huynh T, Miranda K, Tsirigos A, McHardy A, Platt D. Short blocks from the noncoding parts of the human genome have instances within nearly all known genes and relate to biological processes. Proc Natl Acad Sci U S A. 2006;103:6605–6610. doi: 10.1073/pnas.0601688103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 47.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaucheret H, Mallory AC, Bartel DP. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 52.Fischer SE, Butler MD, Pan Q, Ruvkun G. Trans-splicing in C. elegans generates the negative RNAi regulator ERI-6/7. Nature. 2008;455:491–496. doi: 10.1038/nature07274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lau NC, Ohsumi T, Borowsky M, Kingston RE, Blower MD. Systematic and single analysis of Xenopus Piwi-interacting RNAs and Xiwi. EMBO J. 2009;28:2945–2958. doi: 10.1038/emboj.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lau NC. Analysis of small endogenous RNAs Chapter 26. Curr Protoc Mol Biol. 2008;(Unit26):27. doi: 10.1002/0471142727.mb2607s81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.