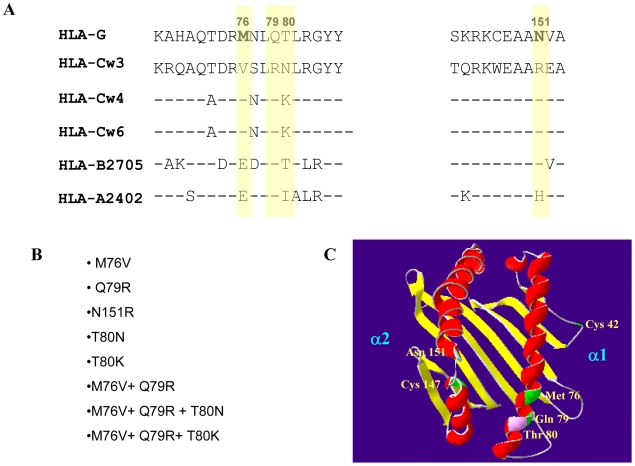
Figure 1. HLA-G is markedly different from HLA-C and other selected MHC class I molecules in the contact residues between KIR and HLA-C.
(A) Sequence alignment between representative MHC class I molecules of the HLA-A, B and C sub-classes and HLA-G in the binding interface with KIR inhibitory receptors. The HLA-G residues that were selected for site-directed mutagenesis are shown in bold and highlighted in yellow. The sequences are shown for two regions (positions 68–85 and 143–153). Conserved residues are indicated by dashes. (B) A ribbon diagram of the crystal structure of HLA-G with the contact residues superimposed. Cys 42 and Cys 147 which form disulfide bridges for the formation of HLA-G complexes and the contact residues that were mutated are indicated. Domains α1 and α2 are also indicated. This backbone modeling of the HLA-G molecule was generated using Swiss-PDB viewer v3.7 software. (C) A list of the single, double and triple mutations which were performed in the HLA-G molecule.