Figure 1.
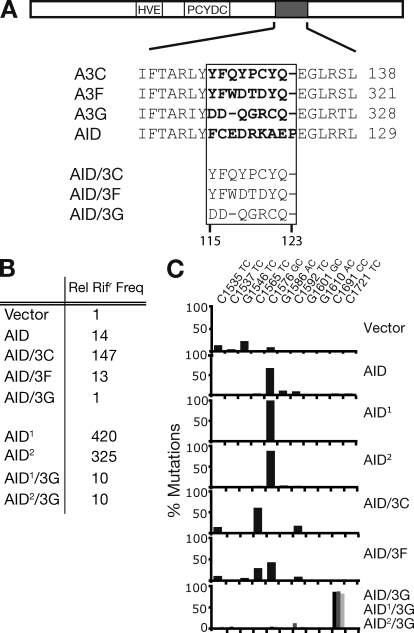
Changing the target specificity of AID. (A) Depiction of AID cDNA showing the Zn coordination domain (HVE and PCYDC), and the region containing a putative substrate contact loop (gray), with alignment to equivalent regions of APOBEC3C, 3F, and 3G shown below. Residues predicted to be in the substrate contact loop are highlighted in bold. In the AID chimeras, residues 115–123 of AID were replaced by equivalent residues in APOBEC3C/F/G as indicated. (B) Bacterial mutator activity of variant AIDs was determined by the mean frequency from 12 independent cultures with which they yielded colonies resistant to rifampicin (Rifr), expressed relative to that given by the vector-only control. AID1 and AID2 are previously described upmutants of wild-type AID (AID1: K10E/T82I/E156G; AID2: K34E/E156G/R157T; Wang et al., 2009). (C) Target specificity of the various AID-derived deaminases as judged by the distribution of rpoB mutations in rifr resistant colonies. Transition mutations at any one of eleven C:G pairs within rpoB can give rise to Rifr. Mutations at a specific C:G pair are expressed as a percentage of the total number of Rifr colonies scored for each deaminase. Results with AID/3G are shaded black, AID1/3G in dark gray, and AID2/3G in light gray (all largely target C1691).