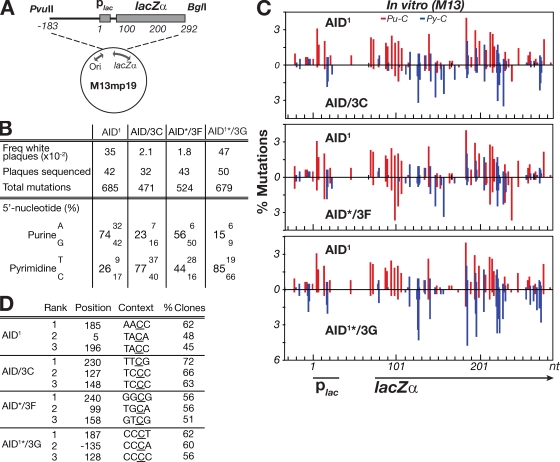
Figure 2.
Mutation spectrum of modified AIDs assayed in vitro on a gapped duplex lacZ target. (A) Depiction of the M13mp19lacZ gapped duplex substrate DNA. (B) 5′-flanking nucleotide preferences of the C mutations produced by the variant AID deaminases. The spectra shown for the AID/3G chimera is from an AID1*/3G derivative in which the asterisk denotes that the protein has been truncated at amino acid position 190 of AID (removing the nuclear export sequence) and is shown to allow comparison with the same AID variant analyzed in transfected DT40 B cells (see Fig. 3). The C-terminal truncations do not detectably alter the patterns of in vitro mutational targeting. (C) Distribution of mutations over a 310-nt stretch of the single-stranded lacZ target. The numbers of independent mutations at each nucleotide position are expressed as a percentage of the total mutation database (as analyzed over the entire 475-nt single-stranded target). Nucleotide position 1 is defined as the start of the lac promoter. Mutations at C residues flanked by a 5′-purine (Pu-C) are shown in red, those flanked by a 5′-pyrimidine (Py-C) in blue. (D) Identity of the three most frequently targeted residues by each deaminase, with targeting expressed as the percentage of clones analyzed in which the relevant cytosine (underlined) was mutated.