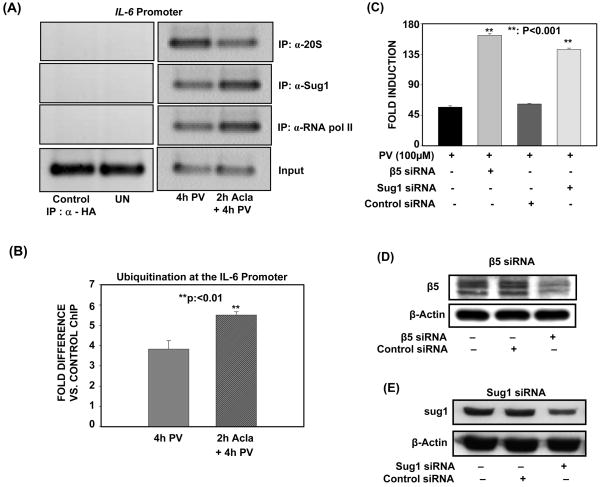
Figure 6. Catalytic core and regulatory subunits of the proteasome are recruited to the IL-6 promoter to negatively regulate PV-induced IL-6 transcription.
(A). ILU-18 cells were either untreated or treated with 100μM PV for 4h, with or without pretreatment with 0.25μM Acla for 2 hours. ChIP assays employing anti-20S, anti-Sug1, and anti-RNA pol II were then performed using immunoprecipitated DNA amplified with primers specific for the IL-6 promoter. ChIP assay employing α-HA (irrelevant antibody) in cells treated with Acla+PV served as a specificity control.
(B). ILU-18 cells were either untreated or treated with 100μM PV for 4h, with or without pretreatment with 0.25μM Acla for 2 hours. ChIP assay employing Anti-Ubiquitin (FK2) was then performed. Immunoprecipitated DNA was analyzed by quantitative real-time PCR analysis using primers amplifying the promoter region of IL-6. Fold difference is Anti-Ubiquitin (FK2) ChIP relative to control ChIP (immunoprecipitated DNA from untreated cells). Statistically significant differences are denoted by **.
(C). ILU-18 cells were either left untreated or transfected twice in 72 hours with either scrambled, β5-specific, or Sug1-specific siRNA. Cells were treated with 100μM PV for 18 hours, following transfection. Fold induction represents normalization of treatment-induced luciferase activity to basal luciferase activity obtained in untreated cells. Data obtained from at least four replicates are presented as mean ± standard error. Statistically significant differences are denoted by **.
(D), (E). ILU-18 cells were either left untreated or transfected twice in 72 hours with either scrambled, β5-specific, or Sug1-specific siRNA. Cells were lysed following transfection and 30μg of each sample was analyzed by immunoblotting using an antibody specific to β5 (D) or Sug1 (E). After stripping, the blot was reprobed with anti-β-actin to demonstrate equal loading.