Abstract
Phytochelatins (PCs) play an important role in detoxification of heavy metals in plants. PCs are synthesized from glutathione by phytochelatin synthase (PCS), a dipeptidyltransferase. Sesbania rostrata is a tropical legume plant that can tolerate high concentrations of Cd and Zn. In this study, the S. rostrata PCS gene (SrPCS) and cDNAs were isolated and characterized. Southern blot and sequence analysis revealed that a single copy of the SrPCS gene occurs in the S. rostrata genome, and produces four different SrPCS mRNAs and proteins, SrPCS1–SrPCS4, by alternative splicing of the SrPCS pre-mRNA. The SrPCS1 and SrPCS3 proteins conferred Cd tolerance when expressed in yeast cells, whereas the SrPCS2 and SrPCS4 proteins, which lack the catalytic triad and the N-terminal domains, did not. These results suggested that SrPCS1 and SrPCS3 have potential applications in genetic engineering of plants for enhancing heavy metal tolerance and phytoremediation of contaminated soils.
Keywords: alternative splicing, heavy metal tolerance, phytochelatin, phytochelatin synthase, phytoremediation, Sesbania rostrata
1. Introduction
Phytochelatins (PCs) have been found in all plant species investigated, a few fungal species and some marine diatoms [1]. PCs can act as high affinity metal chelators and facilitate the vacuolar sequestration of heavy metals, especially Cd2+ ions, and hence play in the plant kingdom an important role in detoxification of some heavy metals [2,3]. PCs have the general structure (γ-Glu-Cys)n-Gly, where n has been reported to be as high as 11, but generally ranges from 2 to 5 [4]; they are synthesized posttranslationally in the presence of heavy metal ions from glutathione (GSH) by PC synthase (PCS), a γ-glutamyl-cysteine transpeptidase [5]. So far, four different models for the catalytic mechanism of PCS have been proposed [5–10], although a consistent model that clearly demonstrates how PCS is activated and how PCs are synthesized has not been established. Alignment of PCS protein sequences reveals a high degree of similarity in the N-terminal domain, whereas the C-terminal domain which turns out to be extremely variable, is missing in the prokaryotic PCS proteins [11,12]. The N-terminal core domain of PCS, which can be referred to the catalytic domain sufficient to confer a PCS activity, has been confirmed by limited proteolysis and mutant analyses [6,8,11,13]. The variable C-terminal region was shown to ensure higher PCS activity, enhance protein stability and respond to a broader spectrum of heavy metals [11]. The stereostructure of the PCS from the cyanobacterium Nostoc sp. PCC7120 (NsPCS) in its native and γ-Glu-Cys-acylated condition was reported [14]. This study represented the first crystal structure of a PCS and established that prokaryotic NsPCS and the eukaryotic Arabidopsis thaliana PC synthase1 (AtPCS1) belong to the papain superfamily of cysteine proteases, with a conserved catalytic machinery that had been deduced from kinetic, protein chemical, and site mutagenic studies of the prototypical AtPCS1 [8,10,15].
PCS genes have been cloned and characterized in Arabidopsis thaliana (AtPCS1, [3,16]), Schizosaccharomyces pompe (SpPCS, [17]), Triticum aestivum (TaPCS1, [6]), Lotus japonicus (LjPCS, [18–20], the model nematode Caenorhabditis elegans (CePCS1, [21,22]) and the prokaryote Nostoc sp. PCC7120 (NsPCS, [8,12]). It has been proposed that PCS is a constitutive enzyme in general and its gene expression is not induced in response to heavy metal exposure [23]. This proposal was supported by the analysis of AtPCS1 expression, which showed that the level of AtPCS1 mRNA was unchanged on exposure to heavy metals such as Cd, Zn, or Cu, or to oxidative stress, salt stress, jasmonic acid or salicylic acid [6,10,24]. During the early stages of plant development, however, the steady-state level of AtPCS1 mRNA increased by 2-fold in 5-day-old Cd-treated Arabidopsis compared to non-treated seedlings [25]. Clemens et al. [17] found that the level of TaPCS1 in wheat roots treated with 100 μM Cd2+ was 5- to 10-fold higher than those of untreated roots. These studies indicated that in specific organs and developmental stages of plants, in which a strong defense against exposure to toxic heavy metals is required, the expression of the PCS genes may be regulated in response to heavy metals.
Sesbania rostrata, a legume native to tropical West Africa, is an annual species that bears stem nodules as well as root nodules associated with Azorhizobium caulinodans, a specialized fast-growing strain of Rhizobia that can fix nitrogen in its free-living form [26]. Yang et al. [27] found that S. rostrata can tolerate some heavy metals, especially Zn and Cd. To understand the mechanism of heavy metal tolerance in S. rostrata, in this study the corresponding PCS gene and its cDNAs were isolated and characterized.
2. Results and Discussion
2.1. Cloning and Sequence Analysis of PCS cDNAs and Gene from S. rostrata
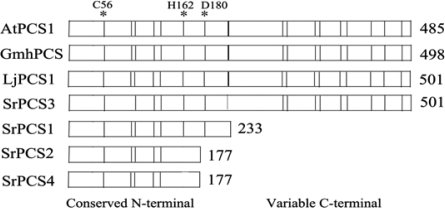
Based on the conserved sequence of the known plant PCS proteins, four full-length cDNA clones encoding PCS in S. rostrata, designated SrPCS1 (GenBank accession no. DQ010916), SrPCS2 (GenBank accession no. GQ204308), SrPCS3 (GenBank accession no. GQ204309) and SrPCS4 (GenBank accession no. GQ204310), were isolated by RT-PCR amplification of conserved cDNA sequences combined with 3′ and 5′-rapid amplification of cDNA ends (RACE). The calculated molecular masses from the deduced amino acid sequences of SrPCS1, SrPCS2, SrPCS3 and SrPCS4 are 26 kD, 19.8 kD, 55.8 kD, and 19.9 kD, respectively (Figure 1).
Figure 1.
Schematic comparison of PCS polypeptides from different plants.
The positions of the conserved Cys residues are indicated by vertical bars. They exhibit at least 60% identical amino acids in pair-wise comparison. The triad Cys-56, His-162 and Asp-180 are indicated as asterisks. AtPCS1, PCS from A. thaliana, (GenBank accession no. AAD41794); LjPCS1, PCS from Lotus japonicus (GenBank accession no. AY633847); SrPCS1-4, PCS1-4 from S. rostrata (GenBank accession nos. DQ010916, GQ204308, GQ204309 and GQ204310).
Among the four SrPCS cDNAs, the ORF of SrPCS3 is the longest one; its amino acid sequence shares 65% identity to Arabidopsis AtPCS1 [16], 87% identity to soybean GmhPCS1 [7] and 87% identity to L. japonicus LjPCS1 [18]. The identity of SrPCS3 with SrPCS1, SrPCS2 and SrPCS4 is 97.9%, 87.6% and 88.1%, respectively. The N-terminal domains of SrPCS3 and SrPC1, but not SrPCS2 and SrPCS4, contain a catalytic triad of Cys-56, His-162 and Asp-180 (Figure 1), which is essential for the activity [9,10,13,14].
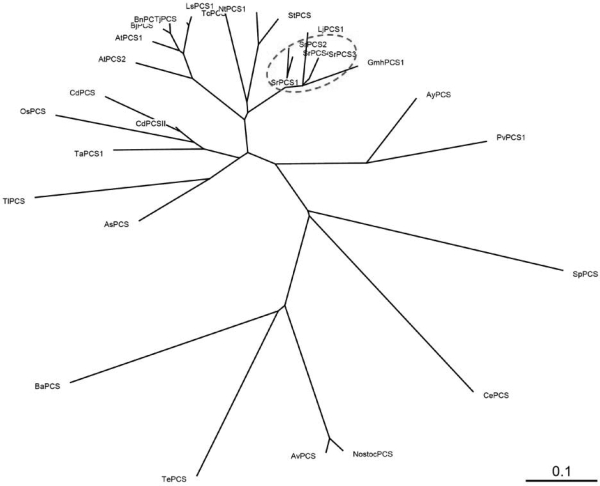
An unrooted phylogenetic tree (Figure 2) was constructed including the majority of the predicted PCS sequences available in the databases. The tree revealed separate clusters, paralleling taxonomic distance, for the PCSs of cyanobacteria, nematodes, yeast, ferns, and the families of vascular plants Poaceae, Allaceae-Typhaceae, Solanaceae, Brassicaceae, and Leguminosae. As shown in Figure 2, SrPCS proteins (in red dashed ellipse of Figure 2) are highly homologous to leguminous soybean GmPCS [7] and Lotus LjPCS1 [18]. The structure and function of PCS genes have only been studied in detail in the model plant Arabidopsis. Two functional PCS genes, AtPCS1 and AtPCS2, which are localized in different chromosomes (At5g44070 and Atlg03980), were found in Arabidopsis [6,10,17,24]. In order to study the structure of S. rostrata PCS gene (SrPCS), we cloned the genomic sequence of SrPCS from the genomic DNA of S. rostrata by genome walking [28] with primers based on the conserved sequence of SrPCS1-4 cDNAs. Unexpectedly, only a 5,112-bp genomic DNA fragment of SrPCS (GenBank accession no. GQ204307) was obtained.
Figure 2.
Phylogenetic analysis of PCS proteins from cyanobacteria, nematodes, fungi, and plants.
The unrooted tree was constructed using the neighbor-joining method (ClustalW) with 1,000 bootstrp replicates. Branch lengths are proportional to genetic distance, which is indicated by a bar (0.1 substitutions per site). Abbreviations and GenBank accession numbers are as follows (in parentheses) AsPCS (Allium sativum AAO13809), AtPCS1 (A. thaliana, AAD41794), AtPCS2 (A. thaliana, AAK94671), AyPCS (Athyrium yokoscense, BAB64932), BjPCS (Brassica juncea, CAC37692), NtPCS1 (Nicotiana tabacum, AAO74500), TaPCS (Triticum aestivum, AAD50592), OsPCS (Oryza sativa, AAO13349), TjPCS (Thlaspi japonicum, BAB93119), TlPCS (Typha latifolia, AAG22095), SpPCS (Schizosaccharomyces pombe, CAA92263), StPCS (Solanum tuberosum, CAD68109), CdPCS (Cynodon dactylon, AAO13810), GmPCS (Glycine max, AAL78384), LjPCS1 (Lotus japonicus, AY633847), NsPCS (Nostoc sp. PCC 7120, NP-485018), SrPCS1 (S. rostrata, DQ010916), SrPCS2 (S. rostrata, GQ204308), SrPCS3 (S. rostrata, GQ204309), SrPCS4 (S. rostrata, GQ204310), PvPCS1 (Pteris vittata, AAT11885), CdPCSII (Cynodon dactylon, AAS48642), AvPCS (Anabaena variabilis ATCC 29413, YP_323464), TePCS (Trichodesmium erythraeum IMS101, YP_722155), BmPCS (Burkholderia mallei SAVP1, YP_993352), BnPCS (Brassica napus, CAK24968), TcPCS (Thlaspi caerulescens, BAB93120), and LsPCS1 (Lactuca sativa, AAU93349).
2.2. Genomic Organization and Pre-mRNA Splicing of SrPCS Gene
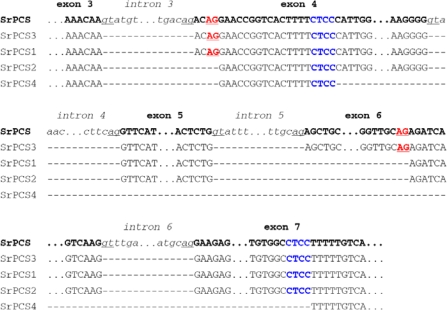
Alignment of the genomic sequence of SrPCS with those of four cDNA sequences of SrPCS1-4 revealed that all four mRNAs (SrPCS1-SrPCS4) were produced from the one SrPCS gene through alternative splicing (Figure 3).
Figure 3.
Alternative splicing of the pre-mRNA for the SrPCS gene.
Exon nucleotides are shown as bold upper-case letters, and intron nucleotides are shown as italic lower-case letters. Canonical GT-AG junctions are underlined. Alternative AG acceptor splice sites are shown as bold upper-case red letters. Atypical splicing sites are shown as bold upper-case blue letters.
By comparing with the sequence of the canonically spliced form SrPCS3 mRNA, eight exons and seven introns were found in SrPCS gene. As shown in Figure 3, SrPCS1 and SrPCS2 are generated through alternative AG acceptor splice sites. And the SrPCS4 is produced by atypical splice mechanism employing direct repeats at splice sites (CTCC, Figure 3), which is similar to the atypical mRNA processing of p53 transcripts in the human brain [29].
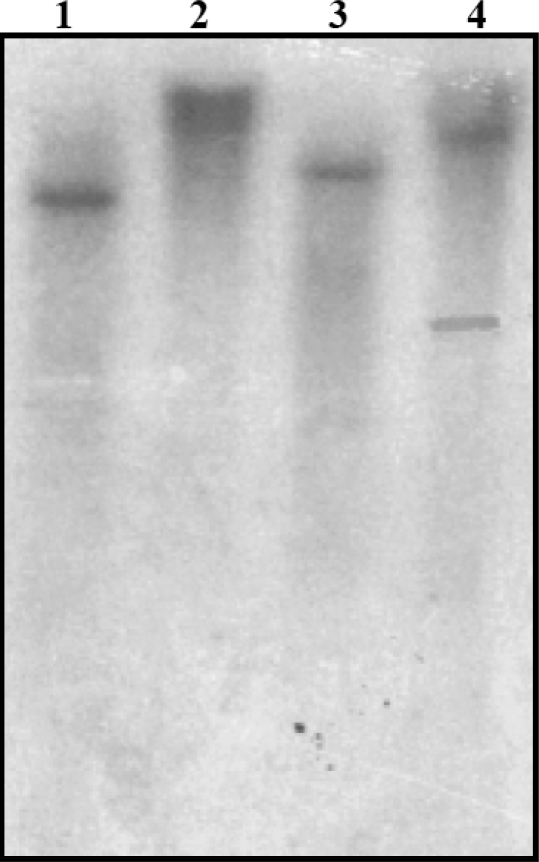
The presence of a single copy of the SrPCS gene in S. rostrata genome was further confirmed by Southern blot analysis (Figure 4). Only a single hybridizing band was detected in DNA samples digested with either EcoRI, BamHI or XbaI, which do not cut within the probe region (Figure 4, lane 1–3). When the genomic DNA was digested with PstI that cut once within the probe region, two hybridizing bands were observed (Figure 4, lane 4). These results indicate that the S. rostrata genome contains only a single copy of the SrPCS gene.
Figure 4.
Southern blot analysis of S. rostrata genomic DNA.
S. rostrata genomic DNA (25μg) digested with EcoRI (lane 1), BamHI (lane 2), XbaI (lane 3), and PstI (lane 4) was separated by agarose gel electrophoresis, blotted onto Hybond-N membrane and hybridized to the 32P-labeled conserved SrPCS cDNA fragment (nt 47-919 of GenBank GQ204309).
2.3. Expression Analysis of SrPCS Gene
Alternative splicing of pre-mRNA is often regulated by specific factors in response to developmental or environmental cues [30–32]. To find out whether the alternative splicing of SrPCS pre-mRNA is regulated by heavy metals, we investigated SrPCS expression in plants in the presence and absence of Cd2+. As in wheat [17] and Brassica juncea [33] plants, SrPCS1–SrPCS4 mRNA expression was very low and could not be detected by RNA blot analysis of total RNA from different organs of S. rostrata. Therefore a more sensitive real time-PCR method was used to estimate the abundance of SrPCS transcripts. Unfortunately, because of very high sequence similarity of the four SrPCS mRNA, only specific primers for SrPCS3 can be designed for real time-PCR analysis.
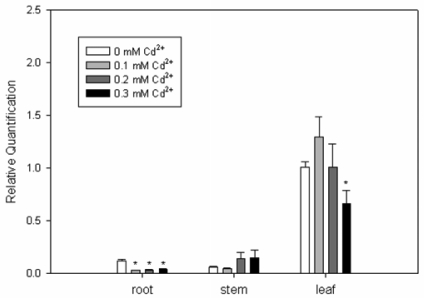
Unlike in Arabidopsis and Brassica juncea, where higher amounts of Arabidopsis AtPCS1 transcript [6] and B. juncea BjPCS1 protein [33] were detected in roots than in leaves, SrPCS3 is more highly expressed in the leaves than roots and stems under normal growth conditions without Cd2+ treatment (Figure 5). SrPCS3 expression was significantly reduced in roots after exposure to different concentration of Cd2+ for 24 hours. No significant change of SrPCS3 expression was observed in stems, whereas SrPCS3 expression was also significantly down-regulated in leaves by 0.3 mM Cd2+ treatment. These observations are contrary to TaPCS1 from wheat [17] and LjPCSs from the model legume Lotus japonicus [18]. Clemens et al. [17] found that TaPCS1 showed a 5- to 10-fold higher expression in wheat roots treated with 0.1 mM Cd2+ for 6 h. Ramos et al. [18] investigated the expression of three PCS genes in roots of L. japonicus, and found that the mRNA expression of LjPCS1-8R increased only slightly (2.1-fold) after 6 h of 0.1 mM Cd2+ treatment, whereas the mRNA levels of LjPC2-7R and LjPCS3-7N increased 2.3- to 2.9-fold and 2.5- to 3.5-fold between 6 and 96 h, respectively.
Figure 5.
Expression analysis of SrPCS3 transcript by real-time PCR.
The relative expression of SrPCS3 to an internal control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in S. rostrata plants, treated with different concentration of Cd2+ for 24 hours, was analyzed by real-time RT-PCR. Mean values plus standard errors are given. * indicates significance at the 5% level.
2.4. Functionality of Four SrPCS Isoforms SrPCS1–SrPCS4
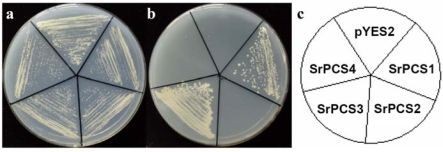
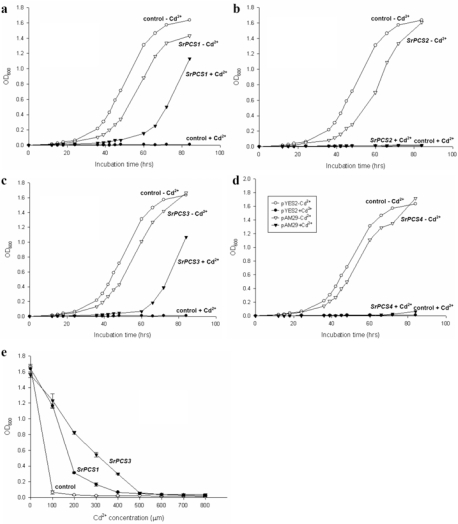
To test the functionality of four SrPCS isoforms, the full-length cDNAs of SrPCS1–SrPCS4 were expressed in S. cerevisiae under the control of the inducible pGAL1 promoter. Cd tolerance assay showed that S. cerevisiae cells expressing SrPCS1 or SrPCS3, but not SrPCS2 and SrPCS4, exhibited strong tolerance to Cd2+ (600 μM), which completely inhibited the growth of cells transformed with the empty pYES2 plasmid or cells expressing SrPCS2 or SrPCS4 (Figure 6). Growth curve experiments (Figure 7, a–d) also demonstrated that expression of SrPCS1 and SrPCS3 conferred enhanced Cd tolerance to yeast cells (Figure 7, a and c), whereas the expression of SrPCS2 and SrPCS4 did not make yeast cells more Cd tolerant (Figure 7, b and d).
Figure 6.
Cd tolerance assay of yeast cells expressing SrPCS cDNAs.
Growth of control S. cerevisiae cells (carrying the empty pYES2 plasmid) and cells expressing one of the SrPCS1–SrPCS4 on YNB plate without (a) and with 600 μM Cd2+ (b). (c) a diagram showing the cells on plates shown in (a) and (b).
Figure 7.
Growth curves of yeast cells expressing SrPCS cDNAs.
Growth of control S. cerevisiae cells carrying the empty pYES2 plasmid and cells expressing SrPCS1 (a), SrPCS2 (b), SrPCS3 (c), or SrPCS4 (d) in YNB medium (supplemented with 1% raffinose + 1% galactose) containing either no (open symbols) or 200 μM Cd2+ (filled symbols). (e) growth of control S. cerevisiae cells carrying the empty pYES2 plasmid and cells expressing SrPCS1 or SrPCS3 in YNB medium (supplemented with 1% raffinose + 1% galactose) containing different concentrations of Cd2+. OD600 of cultures after 65 h is shown.
Dose-response analyses showed that SrPCS3-expressing yeast cells tolerated higher Cd2+ concentrations than the SrPCS1 expressing cells and control cells (Figure 7, e), which may result from the lack of the C-domain that has been shown to ensure higher PCS activity [11]. Alternative splicing, unlike gene promoter activity that primarily regulates the amount of transcripts, changes the structure of transcripts and their encoding proteins [34]. Therefore alternative splicing is one of the most important mechanisms to generate a large number of mRNAs and protein isoforms from a single or few genes [35]. Several studies have shown that about 20% of plant genes have one or more alternative transcript [36–39]. And based on the alignments of ESTs from other legume species against the Medicago truncatula genome sequence, hundreds of legume genes were recently predicted to be alternatively spliced [39].
In this study, we found that a single copy of the SrPCS gene occurs in S. rostrata genome, and that alternative splicing of the SrPCS pre-mRNA produced four different SrPCS mRNAs and proteins, SrPCS1–SrPCS4 (Figure 1). The SrPCS1 and SrPCS3 proteins, whose N-terminal domains contain a catalytic triad of Cys-56, His-162 and Asp-180 (Figure 1), conferred Cd tolerance when expressed in yeast cells (Figure 6 and 7), whereas the SrPCS2 and SrPCS4 proteins, which contain only a Cys-56 of the catalytic triad and lack the N-terminal domains (Figure 1), did not render yeast cells more Cd tolerant (Figure 6 and 7). This observation is similar to the findings of Ramos et al. [18], who showed that a PCS gene (LjPCS2) in model legume Lotus japonicus can be alternatively spliced to encode two protein isoforms, one (LjPCS2-7N) conferring enhanced Cd tolerance to yeast cells while the other (LjPCS2-7R) did not. SrPCS1 and SrPCS3 characterized in this study have potential applications in genetic engineering of plants for enhancing heavy metal tolerance and phytoremediation of contaminated soils [40]. Although we detected the mRNA expression of the four SrPCS isoforms in the current study, the corresponding proteins of these mRNAs and their physiological functions remain to be characterized. In particular, the presence of SrPCS2 and SrPCS4 proteins, which lack the catalytic triad and the N-terminal domains and do not confer enhanced Cd tolerance to yeast cells, needs to be determined in S. rostrata plants, since they are small variants of the full-length protein (about one-third of the SrPCS3) and may not be stable if translated in plant cells.
3. Experimental Section
3.1. Plant Materials
Seed testae of Sesbania rostrata were scalpelled and surface-sterilized in 70% alcohol for 1 minute, followed by three rinses with sterile distilled water, and then soaked in 10% sodium hypochlorite for 15 minutes with gentle agitation. After washing three times with sterile distilled water, the seeds were germinated on 1/2 MS medium [41] for 6 days. These seedlings were then transferred to 1/8 liquid MS without sugar to grow in this medium for 15 days, changing medium every three days, and then transferred to 1/4 liquid MS containing different concentration of cadmium for 1–2 days as needed at 24 ± 2 °C under fluorescent light with a 16-h photoperiod.
3.2. Cloning of PCS cDNAs and Gene from S. rostrata
Total RNA was extracted from seedlings treated with Cd for 2 days as described above using the Trizol reagent (Invirtogen). The cDNA was synthesized from 2 μg of total RNA that was treated by DNaseI before reverse transcription (RT) with the M-MLV (Progema). RT-PCR amplification of the conserved partial sequence of SrPCS cDNAs was performed with a pair of primers (forward primer ZF168, 5′-GCGTTAAGAGACAATGGCGATG-3′; reverse primer ZF169, 5′-GGGAGATGCTAAG-AAAAGGGTACA-3′) based on the sequence of soybean PCS [7]. The amplification parameters were as follows: 5 min at 95 °C, 30 cycles of 40 s at 95 °C, 40 s at 54 °C and 2 min at 72 °C, 10 min at 72 °C in a 20-μl reaction mixtures. The 5′- and 3′- ends of SrPCS cDNAs were isolated with a modified RACE procedure based on template-switching effect and inverse PCR [42]. Genomic DNA from S. rostrata was extracted by a modified CTAB method [43]. SrPCS genomic clone was obtained using the Universal GenomeWalker Kit (Clontech) according to the manufacturer's protocol. PCR products were cloned into pGEM-T easy (Progema) and sequenced. Sequence identity was calculated by the Ident and Sim program in the Sequence Manipulation Suite (http://www.bioinformatics.org/sms/).
3.3. Real Time RT-PCR of SrPCS3
Real time RT-PCR was used for the quantification of the SrPCS3 expression using the Premix Ex Taq™ (Perfect Real Time, TakaRa) with a pair of SrPCS3 specific primers (forward primer ZF332, 5′-GCTTTATACTCTGAGCTG-3′; reverse primer ZF335, 5′-TTAGATGGCAG-TGATGT-3′). As an internal control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was simultaneously amplified to normalize SrPCS3 expression with a pair of GAPDH specific primers (forward primer ZF371, 5′-GCTGGTGCTGATTATGTT-3′; reverse primer, ZF372 5′-GCTCTGGCTTGTATTCCT-3′). PCR cycling conditions were: 15 s at 95 °C; 40 cycles of 5 s at 96 °C, 20 s at 58 °C and 30 s at 72 °C in a 25-μl reaction mix containing 1x SYBR Green I. Each sample was performed in triplicates at least three times. Amplification products were measured and analyzed with an ABI Prism 7900HT sequence detection system and SDS software (Applied Biosystems).
3.4. Southern Blot Analysis of SrPCS
For Southern blot analysis, 25 μg genomic DNA was digested with restriction enzymes and separated on 0.8% agarose gel and transferred to Hybond-N membrane (Amersham) according to Sambrook and Russell [44]. The conserved SrPCS cDNA fragment (nt 47-919 of GenBank GQ204309) was labeled with [α-32p] dCTP using Random Primer Kit (TakaRa) and used to detect DNA on the membrane as previously described [45].
3.5. Expression of Recombinant SrPCS1-4 in Yeast and Cd Tolerance Assay
Four ORFs of SrPCS1-4 cDNAs were amplified by PCR with a pair of SrPCS-specific primers carrying a BamHI and a KpnI restriction enzyme site (forward primer ZF268, 5′-GGTACCATGGCG-ATGGCGGGGTTGTA-3′; reverse primer ZF269, 5′-ATGGGATCCGGAGATGCTAAGAAAAGG-3′), respectively. After digestion with BamHI and KpnI, each of four PCR fragments containing the ORF of SrPCS1-4 was ligated in the yeast expression vector pYES2 (Invitrogen), resulting in four recombinant vectors (pAM26, pAM27, pAM28, and pAM29) that were used to transform Saccharomyces cerevisiae strain INVSc1 (MATK his3v1 leu2 trp1-289 ura3-62) cells according to the supplier’s protocol (Invitrogen). For the Cd tolerance assay, yeast cells containing the empty vector pYES2 or the SrPCS1-4 cDNA constructs were grown in the yeast nitrogen base (YNB) medium supplemented with the appropriate amino acids, 1% galactose and 1% raffinose containing different concentrations of Cd2+ at 30 °C.
4. Conclusions
Our results demonstrate that S. rostrata, a tropical legume plant tolerant to heavy metals, contains a single copy of the SrPCS gene in its genome, which produces four mRNAs encoding different protein isoforms (SrPCS1–SrPCS4) by alternative splicing of the SrPCS pre-mRNA. When expressed in yeast cells, the SrPCS3 protein containing the catalytic triad and the N-terminal domain rendered yeast tolerant to higher Cd2+ concentrations than the SrPCS1 protein containing the catalytic triad but lacking the N-terminal domain. These results suggested that SrPCS1 and SrPCS3 could be used for the production of heavy metal tolerant transgenic plants. The other two alternatively spliced variants, SrPCS2 and SrPCS4, which lack the catalytic triad and the N-terminal domains and do not confer enhanced Cd tolerance to yeast cells, may play different roles in S. rostrata growth and development, although the presence of these two proteins in plants needs to be confirmed.
Acknowledgments
This work was supported by the Research Fund Program of Guangdong Provincial Key Laboratory of Environmental Pollution Control and Remediation Technology, the 863 Program of China (2001AA649020), and by the Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-YW-Z-0723) to Z. F. Xu.
References
- 1.Cobbett C, Goldsbrough P. Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu. Rev. Plant. Biol. 2002;53:159–182. doi: 10.1146/annurev.arplant.53.100301.135154. [DOI] [PubMed] [Google Scholar]
- 2.Grill E, Winnacker EL, Zenk MH. Phytochelatins: the principal heavy-metal complexing peptides of higher plants. Science. 1985;230:674–676. doi: 10.1126/science.230.4726.674. [DOI] [PubMed] [Google Scholar]
- 3.Tennstedt P, Peisker D, Bottcher C, Trampczynska A, Clemens S. Phytochelatin synthesis is essential for the detoxification of excess zinc and contributes significantly to the accumulation of zinc. Plant Physiol. 2009;149:938–948. doi: 10.1104/pp.108.127472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cobbett CS. Phytochelatins and their roles in heavy metal detoxification. Plant Physiol. 2000;123:825–832. doi: 10.1104/pp.123.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grill E, Loffler S, Winnacker EL, Zenk MH. Phytochelatins, the heavy-metal-binding peptides of plants, are synthesized from glutathione by a specific gamma-glutamylcysteine dipeptidyl transpeptidase (phytochelatin synthase) Proc. Natl. Acad. Sci. USA. 1989;86:6838–6842. doi: 10.1073/pnas.86.18.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ha SB, Smith AP, Howden R, Dietrich WM, Bugg S, O'Connell MJ, Goldsbrough PB, Cobbett CS. Phytochelatin synthase genes from Arabidopsis and the yeast Schizosaccharomyces pombe. Plant Cell. 1999;11:1153–1164. doi: 10.1105/tpc.11.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oven M, Page JE, Zenk MH, Kutchan TM. Molecular characterization of the homophytochelatin synthase of soybean Glycine max: relation to phytochelatin synthase. J. Biol. Chem. 2002;277:4747–4754. doi: 10.1074/jbc.M108254200. [DOI] [PubMed] [Google Scholar]
- 8.Tsuji N, Nishikori S, Iwabe O, Matsumoto S, Shiraki K, Miyasaka H, Takagi M, Miyamoto K, Hirata K. Comparative analysis of the two-step reaction catalyzed by prokaryotic and eukaryotic phytochelatin synthase by an ion-pair liquid chromatography assay. Planta. 2005;222:181–191. doi: 10.1007/s00425-005-1513-9. [DOI] [PubMed] [Google Scholar]
- 9.Vatamaniuk OK, Mari S, Lang A, Chalasani S, Demkiv LO, Rea PA. Phytochelatin synthase, a dipeptidyltransferase that undergoes multisite acylation with gamma-glutamylcysteine during catalysis: stoichiometric and site-directed mutagenic analysis of arabidopsis thaliana PCS1-catalyzed phytochelatin synthesis. J. Biol. Chem. 2004;279:22449–22460. doi: 10.1074/jbc.M313142200. [DOI] [PubMed] [Google Scholar]
- 10.Vatamaniuk OK, Mari S, Lu YP, Rea PA. Mechanism of heavy metal ion activation of phytochelatin (PC) synthase: blocked thiols are sufficient for PC synthase-catalyzed transpeptidation of glutathione and related thiol peptides. J. Biol. Chem. 2000;275:31451–31459. doi: 10.1074/jbc.M002997200. [DOI] [PubMed] [Google Scholar]
- 11.Ruotolo R, Peracchi A, Bolchi A, Infusini G, Amoresano A, Ottonello S. Domain organization of phytochelatin synthase: functional properties of truncated enzyme species identified by limited proteolysis. J. Biol. Chem. 2004;279:14686–14693. doi: 10.1074/jbc.M314325200. [DOI] [PubMed] [Google Scholar]
- 12.Tsuji N, Nishikori S, Iwabe O, Shiraki K, Miyasaka H, Takagi M, Hirata K, Miyamoto K. Characterization of phytochelatin synthase-like protein encoded by alr0975 from a prokaryote, Nostoc sp. PCC 7120. Biochem. Biophys. Res. Commun. 2004;315:751–755. doi: 10.1016/j.bbrc.2004.01.122. [DOI] [PubMed] [Google Scholar]
- 13.Romanyuk ND, Rigden DJ, Vatamaniuk OK, Lang A, Cahoon RE, Jez JM, Rea PA. Mutagenic definition of a papain-like catalytic triad, sufficiency of the N-terminal domain for single-site core catalytic enzyme acylation, and C-terminal domain for augmentative metal activation of a eukaryotic phytochelatin synthase. Plant Physiol. 2006;141:858–869. doi: 10.1104/pp.106.082131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vivares D, Arnoux P, Pignol D. A papain-like enzyme at work: Native and acyl-enzyme intermediate structures in phytochelatin synthesis. Proc. Natl. Acad. Sci. USA. 2005;102:18848–18853. doi: 10.1073/pnas.0505833102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rea PA, Vatamaniuk OK, Rigden DJ. Weeds, worms, and more. Papain's long-lost cousin, phytochelatin synthase. Plant Physiol. 2004;136:2463–2474. doi: 10.1104/pp.104.048579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vatamaniuk OK, Mari S, Lu YP, Rea PA. AtPCS1, a phytochelatin synthase from Arabidopsis: isolation and in vitro reconstitution. Proc. Natl. Acad. Sci. USA. 1999;96:7110–7115. doi: 10.1073/pnas.96.12.7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clemens S, Kim EJ, Neumann D, Schroeder JI. Tolerance to toxic metals by a gene family of phytochelatin synthases from plants and yeast. EMBO J. 1999;18:3325–3333. doi: 10.1093/emboj/18.12.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramos J, Clemente MR, Naya L, Loscos J, Perez-Rontome C, Sato S, Tabata S, Becana M. Phytochelatin synthases of the model legume Lotus japonicus. A small multigene family with differential response to cadmium and alternatively spliced variants. Plant Physiol. 2007;143:1110–1118. doi: 10.1104/pp.106.090894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramos J, Naya L, Gay M, Abian J, Becana M. Functional characterization of an unusual phytochelatin synthase, LjPCS3, of Lotus japonicus. Plant Physiol. 2008;148:536–545. doi: 10.1104/pp.108.121715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loscos J, Naya L, Ramos J, Clemente MR, Matamoros MA, Becana M. A reassessment of substrate specificity and activation of phytochelatin synthases from model plants by physiologically relevant metals. Plant Physiol. 2006;140:1213–1221. doi: 10.1104/pp.105.073635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clemens S, Schroeder JI, Degenkolb T. Caenorhabditis elegans expresses a functional phytochelatin synthase. Eur. J. Biochem. 2001;268:3640–3643. doi: 10.1046/j.1432-1327.2001.02293.x. [DOI] [PubMed] [Google Scholar]
- 22.Vatamaniuk OK, Bucher EA, Ward JT, Rea PA. A new pathway for heavy metal detoxification in animals. Phytochelatin synthase is required for cadmium tolerance in Caenorhabditis elegans. J. Biol. Chem. 2001;276:20817–20820. doi: 10.1074/jbc.C100152200. [DOI] [PubMed] [Google Scholar]
- 23.Grill E, Winnacker EL, Zenk MH. Phytochelatins, a class of heavy-metal-binding peptides from plants, are functionally analogous to metallothioneins. Proc. Natl. Acad. Sci. USA. 1987;84:439–443. doi: 10.1073/pnas.84.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cazale AC, Clemens S. Arabidopsis thaliana expresses a second functional phytochelatin synthase. FEBS Lett. 2001;507:215–219. doi: 10.1016/s0014-5793(01)02976-3. [DOI] [PubMed] [Google Scholar]
- 25.Lee S, Korban SS. Transcriptional regulation of Arabidopsis thaliana phytochelatin synthase (AtPCS1) by cadmium during early stages of plant development. Planta. 2002;215:689–693. doi: 10.1007/s00425-002-0821-6. [DOI] [PubMed] [Google Scholar]
- 26.Somasegaran P, Hoben HJ. Handbook for Rhizobia: methods in legume- Rhizobia technology. Springer-Verlag; New York, NY, USA: 1994. [Google Scholar]
- 27.Yang ZY, Chen FH, Yuan JG, Zheng ZW, Wong MH. Responses of Sesbania rostrata and S. cannabina to Pb, Zn, Cu and Cd toxicities. J. Environ. Sci.-China. 2004;16:670–673. [PubMed] [Google Scholar]
- 28.Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikoshkov A, Hurd YL. p53 splice variants generated by atypical mRNA processing confer complexity of p53 transcripts in the human brain. Biochem. Biophys. Res. Commun. 2006;351:383–386. doi: 10.1016/j.bbrc.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 30.Li L, Howe GA. Alternative splicing of prosystemin pre-mRNA produces two isoforms that are active as signals in the wound response pathway. Plant Mol. Biol. 2001;46:409–419. doi: 10.1023/a:1010645330275. [DOI] [PubMed] [Google Scholar]
- 31.Kazan K. Alternative splicing and proteome diversity in plants: the tip of the iceberg has just emerged. Trends Plant Sci. 2003;8:468–471. doi: 10.1016/j.tplants.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Ali GS, Reddy ASN. Regulation of alternative splicing of pre-mRNAs by stresses. In: Reddy ASN, Golovkin M, editors. Nuclear pre-mRNA Processing in Plants. Volume 326. Springer; Berlin, Heidelberg, Germany: 2008. p. 257. [DOI] [PubMed] [Google Scholar]
- 33.Heiss S, Wachter A, Bogs J, Cobbett C, Rausch T. Phytochelatin synthase (PCS) protein is induced in Brassica juncea leaves after prolonged Cd exposure. J. Exp. Bot. 2003;54:1833–1839. doi: 10.1093/jxb/erg205. [DOI] [PubMed] [Google Scholar]
- 34.Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 35.Ben-Dov C, Hartmann B, Lundgren J, Valcarcel J. Genome-wide analysis of alternative pre-mRNA splicing. J. Biol. Chem. 2008;283:1229. doi: 10.1074/jbc.R700033200. [DOI] [PubMed] [Google Scholar]
- 36.Barbazuk WB, Fu Y, McGinnis KM. Genome-wide analyses of alternative splicing in plants: Opportunities and challenges. Genome Res. 2008;18:1381–1392. doi: 10.1101/gr.053678.106. [DOI] [PubMed] [Google Scholar]
- 37.Wang BB, Brendel V. Genomewide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. USA. 2006;103:7175–7180. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campbell MA, Haas BJ, Hamilton JP, Mount SM, Buell CR. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics. 2006;7:327. doi: 10.1186/1471-2164-7-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang BB, O'Toole M, Brendel V, Young ND. Cross-species EST alignments reveal novel and conserved alternative splicing events in legumes. BMC Plant Biol. 2008;8:17. doi: 10.1186/1471-2229-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raskin I, Ensley BD. Phytoremediation of Toxic Metals: Using Plants to Clean up the Environment. Wiley; New York, NY, USA: 2000. [Google Scholar]
- 41.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962;15:473–497. [Google Scholar]
- 42.Huang JC, Chen F. Simultaneous amplification of 5′ and 3′ cDNA ends based on template-switching effect and inverse PCR. BioTechniques. 2006;40:187–189. doi: 10.2144/000112051. [DOI] [PubMed] [Google Scholar]
- 43.Sharma AD, Gill PK, Singh P. DNA isolation from dry and fresh samples of polysacchariderich plants. Plant Mol. Biol. Rep. 2002;20:415. [Google Scholar]
- 44.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press; New York, NY, USA: 2001. [Google Scholar]
- 45.Xu ZF, Qi WQ, Ouyang XZ, Yeung E, Chye ML. A proteinase inhibitor II of Solanum americanum is expressed in phloem. Plant Mol. Biol. 2001;47:727–738. doi: 10.1023/a:1013623628857. [DOI] [PubMed] [Google Scholar]