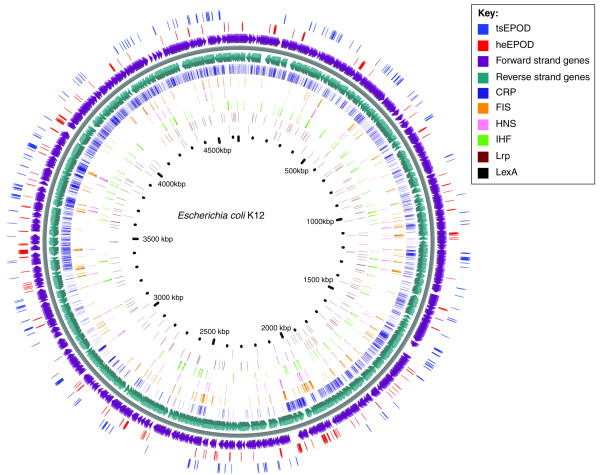
Figure 1.
Occupancy profiles of DNA-binding factors on the Eschericha coli K12 chromosome (gray line), based on integrated results from three different laboratories. Genes are represented by purple and turquoise arrows on forward and reverse strands, respectively. Outer circles represent the EPODs (domains greater than 1 kb with high protein occupancy) reported by Vora et al. [3]. EPODs were clustered using the median expression level across domains obtained from the supplementary material to [3]: heEPODs are red, and tsEPODs are blue. Inner circles represent individual ChIP-chip binding patterns of known transcription factors: CRP [17]; Fis, HN-S, IHF, and Lrp [18]; and LexA [19]. All ChIP-chip data were obtained from the respective supplementary material. The figure was created using the CGview program [24]. CRP, cAMP receptor protein; FIS, factor for inversion stimulation; heEPOD, highly expressed extended protein occupancy domain; H-NS, histone-like nucleoid structuring protein; IHF, integration host factor; Lrp, leucine-responsive protein; LexA, SOS regulatory protein; tsEPOD, transcriptionally silent extended protein occupancy domain.