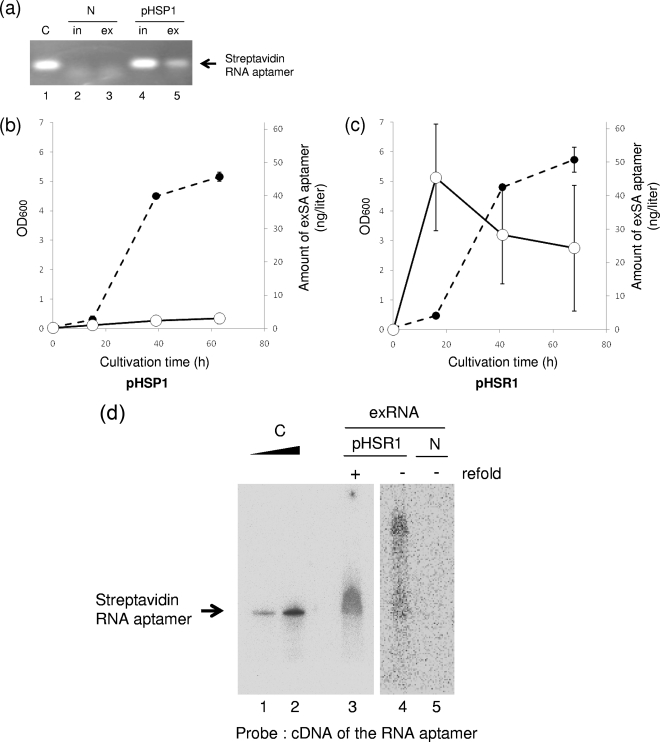
FIG. 3.
Production and release of the streptavidin RNA aptamer into the culture medium. (a) Detection of the streptavidin RNA aptamer sequence. Using streptavidin RNA aptamer-specific primers, the aptamer sequences were amplified by RT-PCR from intracellular (in) and extracellular (ex) RNA preparations from R. sulfidophilum harboring pHSP1 in the early stationary phase of growth. N and pHSP1 represent the RNA preparations from R. sulfidophilum DSM 1374T only (no plasmid) and the bacterium transformed with pHSP1, respectively. C indicates a positive control using the in vitro transcript of the streptavidin RNA aptamer as a template for RT-PCR. (b and c) Extracellular production of the streptavidin RNA aptamer. Time courses of extracellular production of the RNA aptamer from the bacteria harboring pHSP1 (b) and pHSR1 (c) are shown. The amounts of RNA aptamers were measured by quantitative RT-PCR as described in Materials and Methods. Closed circles, cell growth (optical densities at 600 nm [OD600]); open circles, amounts of extracellular streptavidin RNA (exSA) aptamer (ng/liter culture). (d) Northern blotting analysis of the streptavidin RNA aptamer produced. To confirm the size of the RNA, Northern analysis of extracellular-RNA preparations was performed using a cDNA probe for the streptavidin RNA aptamer. C indicates a positive control using the in vitro transcript of the streptavidin RNA aptamer. Clear, sharp bands are shown. This aptamer has a size of 80 nucleotides and can be used as an accurate size marker. N and pHSR1 represent the extracellular RNA preparations (exRNA) from R. sulfidophilum DSM 1374T only (no plasmid) and the bacterium transformed with pHSR1, respectively; refold indicates additional incubation to enhance self-cleavage (see the text).