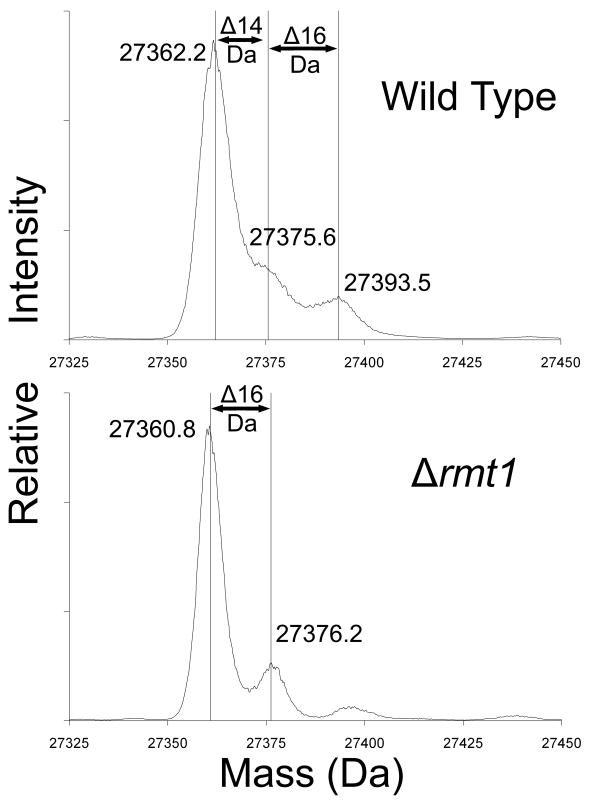
Fig. 3.
Loss of methylation of Rps2 in yeast cells lacking the Rmt1 protein arginine methyltransferase. Small subunits were isolated from the ribosomal fraction of RMT1+ cells (wild type) and of Δrmt1- cells prepared as described in Methods and materials but in 1 l of growth media without the [3H]AdoMet incubation. Due to greater cell pellet volumes, 4.5 ml buffer A was used to lyse each cell pellet and ribosomal pellets were individually re-suspended in 1 ml buffer B. An abbreviated gradient of 7%-27% sucrose made in buffer B was centrifuged for 16 h under the same conditions noted in Materials and methods. Small subunit proteins were prepared as described in Materials and methods except that dialysis was not performed and the sample supernatants after ethanol and acetic acid treatment were directly concentrated by vacuum centrifugation for about 35 min to bring the total volumes to approximately 100 μl. A final centrifugation of 20,000 × g for 5 min removed any particulate matter from solution, and 30 μl of each concentrated sample was analyzed by liquid chromatography/mass spectrometry using an Applied Biosystems Q-Star Elite instrument as described in Materials and methods. Data for the peak at 51 min containing Rps2 were de-convoluted using Analyst software and the resulting average masses of significant peaks are shown. Mass changes are indicated for 14 Da (methylation) as well as 16 Da (oxidation).