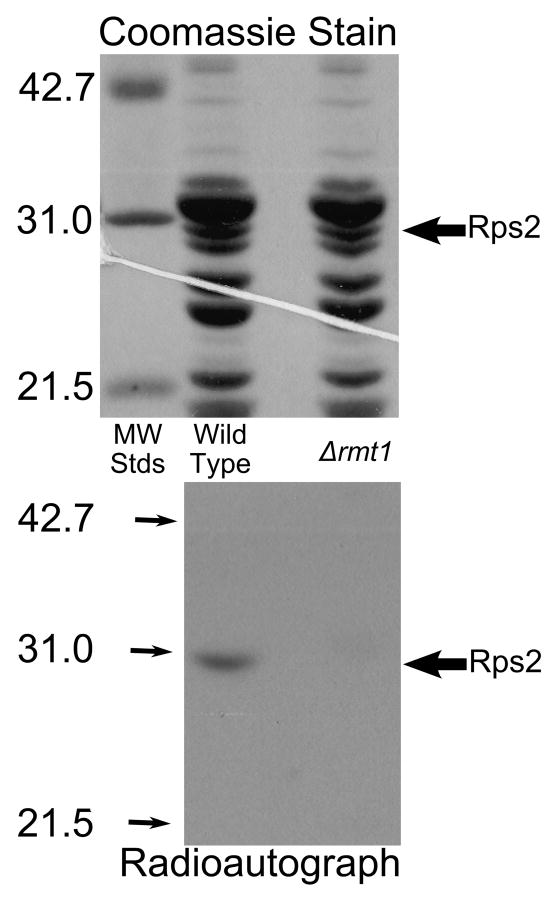
Fig. 4.
Loss of methylation of Rps2 in Δrmt1 mutant cells. Intact cells were labeled with [3H]AdoMet as described in Materials and methods except that cells were grown initially in 1 l of medium and labeled with 2.0 nmol of [3H]AdoMet (Perkin Elmer Life Sciences, 83.3 Ci/mmol) in 40 ml of fresh YPD and the resulting cell pellets were lysed individually using 2 g beads and 4.5 ml buffer A for a total of ten cycles. Small ribosomal subunits were prepared by sucrose gradient centrifugation and the protein fraction isolated by ethanol/acetic acid treatment as described in Materials and methods. 50 μg of small subunit proteins were separated by SDS gel electrophoresis for 35 mA for 40 min and then 25 mA for 5 h with molecular weight standards (Bio-Rad Low Molecular Weight; hen ovalbumin, bovine carbonic anhydrase, and soybean trypsin inhibitor) in an adjoining lane. After protein staining (top panel), the gel was treated in EN3HANCE (Perkin Elmer Life Sciences) for 1 h, followed by a 20 min wash in water before being dried under vacuum for 2 h at 80° C followed by 1 h without heat. The gel was placed on Kodak BIOMAX XAR film for 3 months at -80° C (bottom panel). Large arrows indicate the position of the Rps2 band; small arrows indicate the position of the standards in the radioautograph.