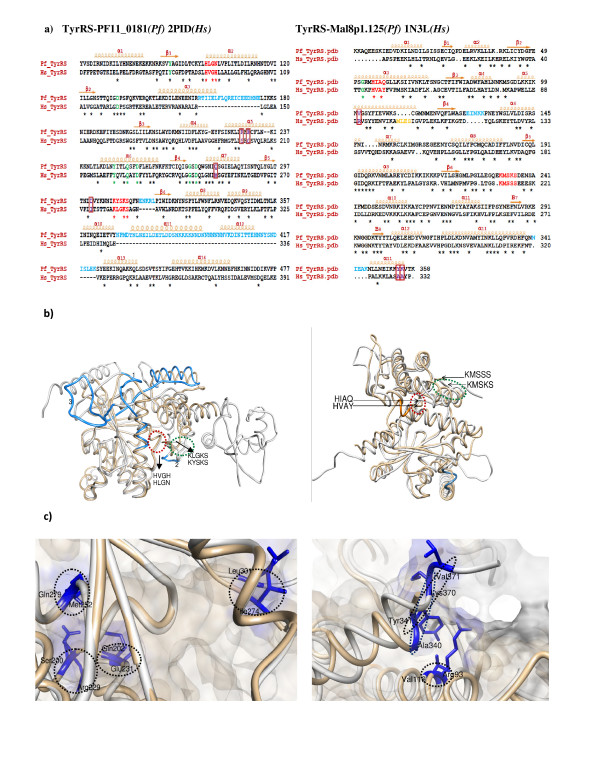
Figure 7.
Left and right panels of the figure represent sequence and structural comparison of bacterial type Plasmodium TyrRS (PF11_0181) with human mitochondrial TyrRS (2PID) and the cytosolic Plasmodium TyrRS (Mal8p1.125) with human cytosolic TyrRS (1N3L). a) A structure-based sequence alignment of the catalytic domain of Plasmodium TyrRSs with human TyrRSs. Insertions in Pf and human sequences are colored in light blue and orange respectively. Class I synthetase conserved motifs are colored red. Residues involved in tRNA recognition and catalysis are indicated in green (same residues in Pf and Hs) and violet & boxed (different in Pf and Hs). The secondary structural elements are shown above the sequence alignments. Conserved residues are indicated by asterisk below the sequence alignment. (b) Superposition of Pf-TyrRS and Hs-TyrRS depicting the structural differences. Pf-Tyr is colored grey and Hs-TyrRS is colored tan. Insertions in Pf-TyrRSs are highlighted in blue whereas Hs-TyrRS insertions are in orange. Motif 1 in Pf (PF11_0181 - HLGN and Mal8p1.125 - HIAQ) and Hs (2PID - HVGH and 1N3L - HVAY) TyrRSs has been encircled red whereas Motif 2 in Pf (PF11_0181 - KLGKS and Mal8p1.125 - KMSKS) and Hs (2PID - KYSKS and 1N3L - KMSSS) is encircled green. (c) Snapshot of the active sites of Pf and Hs TyrRSs (superimposed) structures. Non-conserved active site residues colored violet are encircled.