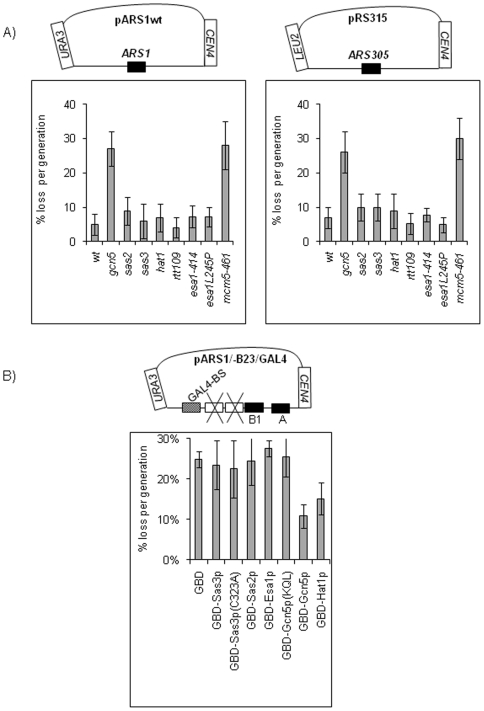
Figure 1. GCN5 regulates mini-chromosome stability.
A. Disruption of GCN5 increases the loss rate of ARS1- or ARS305- mini-chromosomes. Maps of pARS1wt (ARS1, CEN4, URA3) and pRS315 (ARS305, CEN4, LEU2) mini-chromosomes are shown on the top of the graphs. The mini-chromosomes were transformed in the strains shown at the bottom of the graphs. After growth on selective plates (SC-ura or SC-leu, respectively) three colonies were transferred for 20 generations in rich medium (YPD) and plated on YPD and SC-ura or SC-leu, respectively. The proportions of cells with URA+ or LEU+ phenotypes were assessed and the average loss rate per generation was calculated (%) and plotted. Error bars represent standard deviations between the three parallel counts in the experiment. A typical outcome of one of three independent experiments is shown. B. Tethering of Gcn5p increases the stability of pARS1/-B23/GAL4. A diagram of pARS1/-B23/GAL4 (ARS1-B2-B3+GAL4, CEN4, URA3) is shown on the top. The B2 and B3 elements of ARS1 are destroyed and a GAL4 binding site (GAL4-BS) is inserted next to B3. DF5gal4Δ cells were co-transformed with pARS1/-B23/GAL4 and episomes (2 µm/HIS3) expressing the GAL4 DNA binding domain fused to the proteins indicated below the graph. Gcn5p(KQL) lacks HAT activity. Average pARS1/-B23/GAL4 loss per generation and standard deviation were calculated as in Figure 1a. A typical outcome of one of two independent experiments is shown.