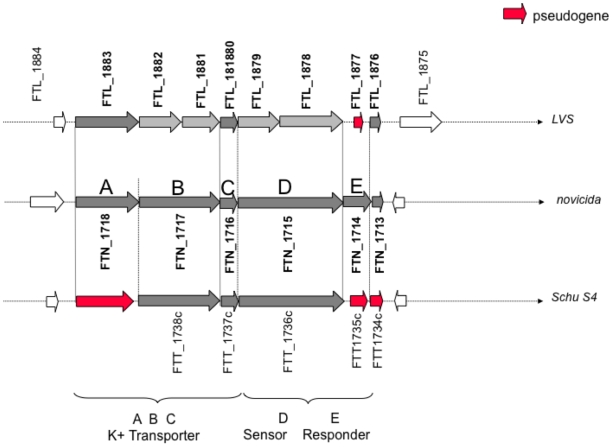
Figure 2. The kdp locus in F. tularensis subsp. holartica (LVS), novicida, and tularensis.
The prototypical Kdp potassium transport system comprises: i) the transporter, composed of KdpA, KdpB and KdpC; and ii) the TCS, regulating its expression, and composed of KdpD, the sensor kinase; and KdpE, the response regulator. Upper line, the LVS locus. Whenever an FTL number has been attributed, it is indicated above the orf. Middle line, F. tularensis subsp. novicida which comprises an intact kdp locus. A, B, D and E, above each gene refer to kdpA, kdpB, kdpC, kdpD and kdpE, respectively. Lower line, F. tularensis subsp. tularensis. In dark grey, the intact genes; in light grey, the interrupted genes (but annotated as genes in the KEGG database); in red, the interrupted genes annotated as pseudogenes (in KEGG). KdpB. In F. tularensis subsp. novicida, the kdpB gene (FTN_1717; 2,040 bp) encodes a 679 aas protein. In LVS, the proximal portion of the gene carries: i) an in frame 63 bp deletion (deletion of nucleotides 509 to 571); and ii) a single nucleotide insertion (between nucleotides 1,096 and 1,097; FTN_1717 numbering), leading to a premature termination of the coding sequence (creation of a TGA stop codon 9 bp downstream of the insertion). The resulting 374 aas truncated protein is designated FTL_1882. An ATG codon (in frame with the rest of the kdpB sequence of FTN_1717) is found immediately downstream of the stop codon (29 bp) of FTL_1882, leading to the prediction of a second open reading frame of 301 aas designated FTL_1881. Blastn analysis of the nucleotide sequence of LVS corresponding to gene FTN_1717 (i.e. 1,978 bp from the start codon of FTL_1882 to the stop codon of FTL_1881) reveals that the entire LVS sequence is 100% identical to F. tularensis subsp. holarctica FTNF002-00 genome region 1,811,308 to 1,809,331 and >99% identical to F. tularensis subsp. holarctica OSU18 genome region 1,815,047 to 1,813,070 (with a unique C to A substitution at position 71 of FTL_1882). Thus, in the three subsp. holarctica genomes available, the proximal part of the kdpB gene carries the same in frame deletion and the gene is interrupted by the same single nucleotide insertion. KdpD. In F. tularensis subsp. novicida, the kdpD gene (FTN_1715; 2,682 bp) encodes a 893 aas protein. In LVS, the corresponding gene is interrupted by a single nucleotide insertion (between nucleotides 981 and 982; FTN_1715 numbering), leading to a premature termination of the coding sequence (creation of a TGA stop codon 32 bp downstream of the insertion). The resulting truncated predicted orf is designated FTL_1879 (337 aa). An ATG codon (in frame with the rest of the kdpD sequence of FTN_1715) is found downstream of the stop codon (92 bp) of FTL_1879, leading to the prediction of a second orf of 525 aas, designated FTL_1878. Blastn analysis of the nucleotide sequence of LVS (2,683 bp) corresponding to gene FTN_1715 reveals that the entire LVS sequence is 100% identical to F. tularensis subsp. holarctica OSU18 genome region 1,812,395 to 1,809,713100 and 99% identical to F. tularensis subsp. holarctica FTNF002-00 genome region 1,808,656 to 1,805,974 (with 3 single nucleotide substitutions). Thus, in the three subsp. holarctica genomes available, the kdpD gene is interrupted by the same single nucleotide insertion. KdpE. In F. tularensis subsp. novicida, the kdpE gene (FTN_1714; 687 bp) encodes a 228 aas protein. In LVS, the corresponding region is not predicted to encode a protein (FTL_1877 is designated pseudogene). Comparison of the F. tularensis subsp. novicida and LVS nucleotide sequences reveal the presence of a 13 bp deletion in the proximal portion of the LVS sequence (deletion of nucleotides 45 to 57, FTN_1714 numbering), resulting in a frame shift and the premature termination of the protein sequence (a TGA stop codon is found 11 bp downstream of the deletion). Blastn analysis of the nucleotide sequence of LVS (671 bp) shows a 100% identity with the F. tularensis subsp. holarctica OSU18 genome region 1,809,671 to 1,809,001; and >99% identity with F. tularensis subsp. holarctica FTNF002-00 genome region 1,805,932 to 1,805,262 (1 single nucleotide substitution) as well as with F. tularensis subsp. tularensis Schu S4 genome region 1,823,954 to 1,823, 284 (2 single nucleotide substitution). Thus, in the three subsp. holarctica genomes available and in Schu S4, the kdpE gene is a pseudogene resulting from the same 13 bp deletion. The regions containing the mutations in genes kdpB (FTL_1881-1882), kdpD (FTL_1878-1879) and kdpE (FTL_1877), have been resequenced by sequencing of cloned PCR products. This analysis fully confirmed the published sequence of LVS.