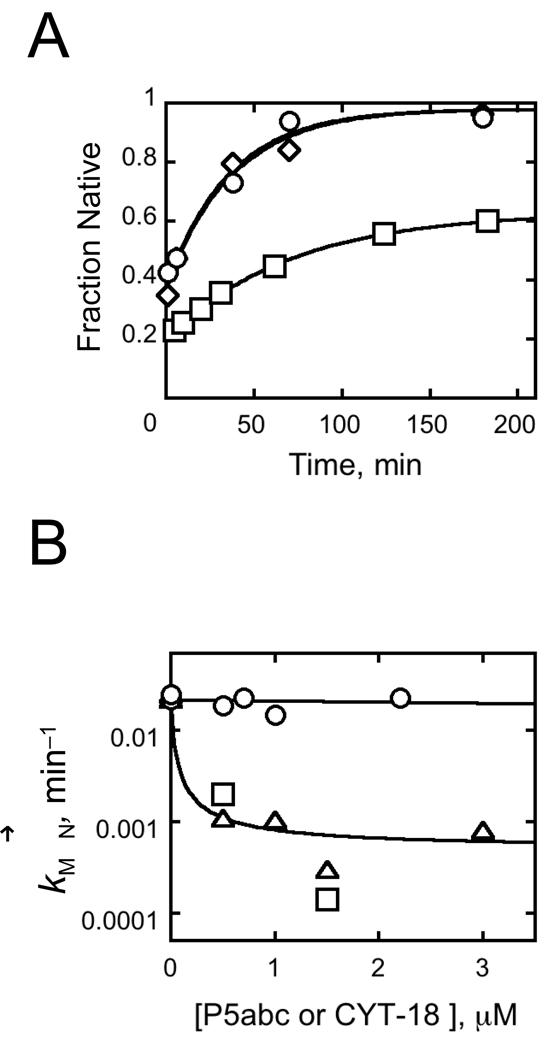
Fig. 10.
CYT-18 does not slow refolding of misfolded ribozyme. A, Time courses of native ribozyme accumulation. After a brief incubation of 200 nM EΔP5abc with 10 mM Mg2+ to allow misfolding, CYT-18 was added (700 nM, circles or 2.2 µM, triangles) and the fraction of native ribozyme was determined by activity at various times thereafter. Values are normalized as described in the Fig. 3 legend. Both reactions gave rate constants of 0.023 min−1. The squares show an equivalent reaction in the absence of CYT-18 (0.015 min−1). B, Refolding rate constants from panel A and additional experiments are plotted against CYT-18 concentration (circles). Results from reactions in which P5abc RNA was added instead of CYT-18 are also shown. Squares show reactions under identical conditions as those with CYT-18 (50 mM Na-MOPS, pH 7.0, 10 mM MgCl2, plus 2.5 mM Tris-Cl, 1% glycerol, and 10 mM KCl), and triangles show reactions under standard RNA folding conditions (50 mM Na-MOPS, pH 7.0, 10 mM Mg2+).