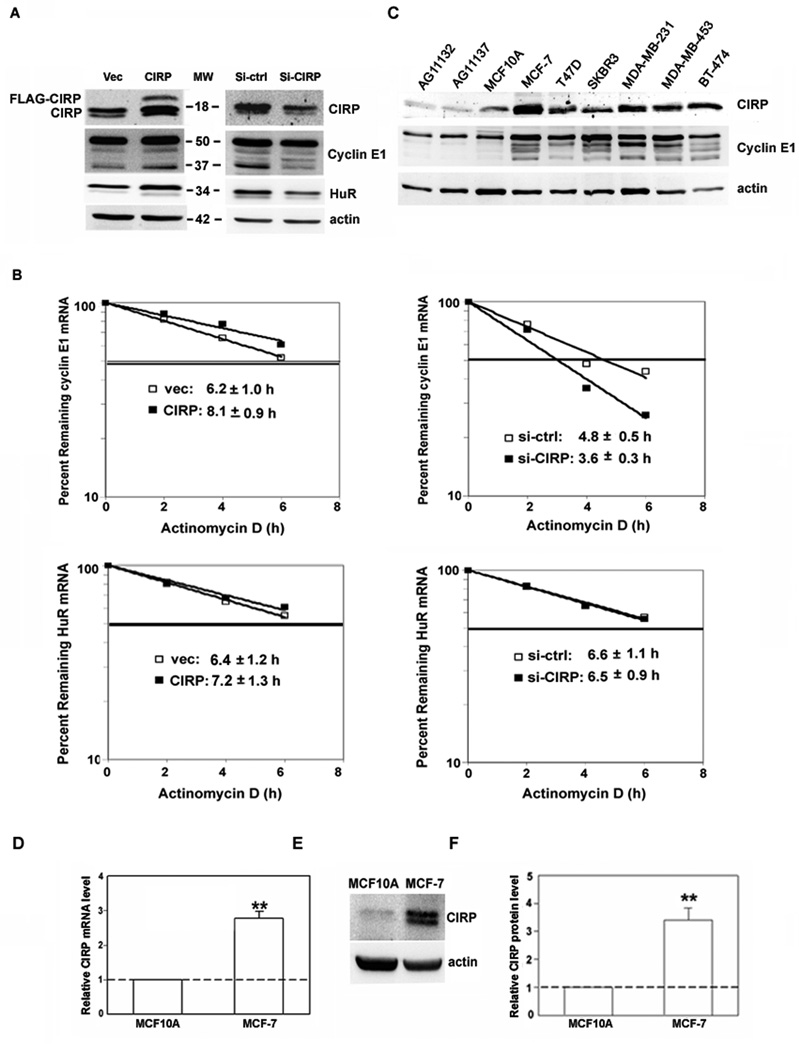
Fig. 2.
Regulation of cyclin E1 and HuR expression by CIRP knockdown and overexpression. A, MCF-7 cells were transfected with CIRP siRNA (Si-CIRP), control siRNA (Si-ctrl), pTracer-CMV2 (Vec), or pTracer-CMV2-CIRP (CIRP). Total protein was extracted 72 hr after transfection and the levels of CIRP, cyclin E1 and HuR were analyzed by western blotting. Equal loading of protein was determined by re-probing for β-actin. B, MCF-7 cells were synchronized at late G1 phase 24h after transfection. Total RNA was extracted at the indicated time after the addition of actinomycin D. Real time RT-PCR was used to analyze HuR and cyclin E1 mRNA level. Data were normalized to GAPDH mRNA and plotted on semi logarithmic scales. p < 0.05 for cyclin E1 mRNA in CIRP/siCIRP cells vs. controls. C, Western blotting for CIRP and cyclin E1 in total protein extracts from normal mammary epithelial cells (lanes 1–2: AG11132, AG11137), immortalized nontumorigenic mammary epithelial cells (lane 3: MCF10A) and 6 breast carcinoma cell lines (lanes 4–9: MCF-7, T47D, SKBR3, MDA-MB-231, MDA-MB-453, BT-474). The blot was stripped and re-probed for β-actin to control for protein loading. D, CIRP mRNA level was analyzed by real-time RT-PCR and expressed as relative level after normalization to GAPDH. E, CIRP protein level was analyzed by western blotting and blots stripped and re-probed for β-actin. F, The relative quantity of CIRP protein was calculated from E after normalization to β-actin. For all experiments, n = 3, **, p < 0.01 versus MCF10A.