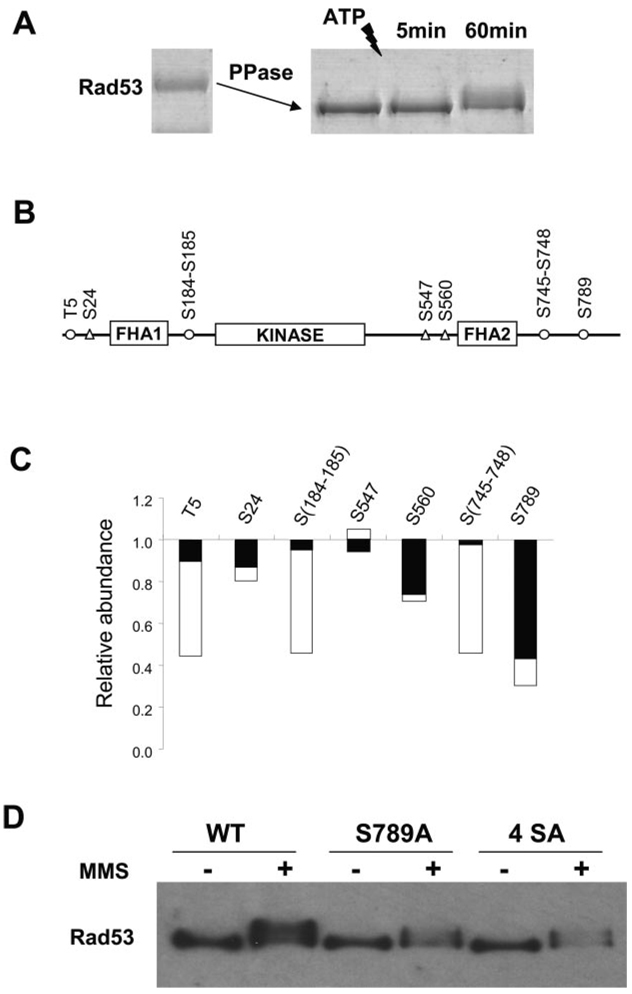
FIG. 5.
A, recombinant Rad53 was first dephosphorylated by λ-phosphatase, and then phosphatase inhibitors were added. Autophosphorylation of Rad53 was initiated by the addition of ATP and stopped after either 5 or 60 min. Tryptic peptides from the dephosphorylated Rad53 protein were labeled by d0-N-isotag, whereas those from 5- or 60-min kinase reaction were labeled by d6-N-isotag and then combined with the d0-N-isotag-labeled sample, respectively. These samples were subjected to IMAC purification and MS analysis for quantitation. B, location of phosphorylation sites identified for Rad53. Unfilled circles indicate the phosphorylation sites after 5 min, whereas triangles indicate the additional phosphorylation sites after 1 h of kinase reaction. C, the depletion of the corresponding unphosphorylated peptides caused by phosphorylation after 5 min (black bar) and 60 min (shaded bar). More detailed information can be found in supplemental Table IV. D, Ser789 appears to locate in a classical bipartite NLS (30), 785KRIHSVSLSQSQIDPSKKVKRAK807, with the basic residues indicated in bold letters. Anti-FLAG Western blot analyses of wild type (WT) and mutants of Rad53-His-FLAG showed that Ser789Ala mutation abolishes the abundance increase of Rad53 after MMS treatment. A quadruple Rad53 4SerAla mutant (Ser789Ala, Ser791Ala, Ser793Ala, and Ser795Ala) showed similar results.