Fig. 1.
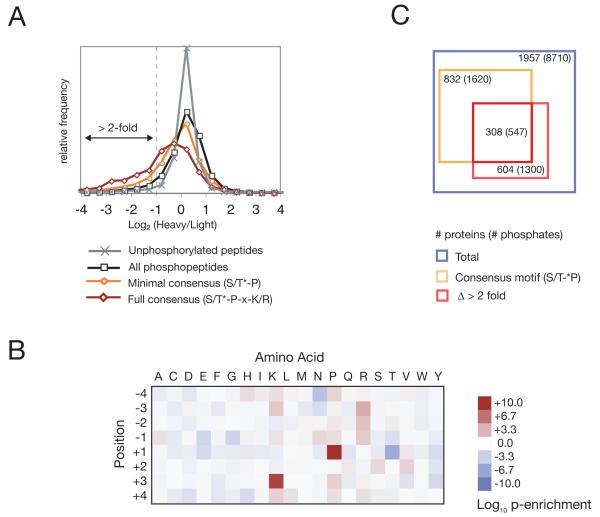
Large-scale identification of Cdk1 substrates in vivo. (A) Distributions of log2 heavy/light (H/L) ratios for unphosphorylated peptides (gray), all phosphopeptides (black) and phosphopeptides containing a minimal (orange) or full (red) Cdk1 consensus phosphorylation motif. (B) Log10 p-values (binomial distribution) for the enrichment (red) or depletion (blue) of each amino acid (columns) at each position flanking the phosphorylated serine or threonine (rows) in phosphopeptides that changed greatly in abundance (log2 H/L < −3) relative to residues flanking serines and threonines proteome-wide. (C) Venn diagram representing the number of proteins and unique phosphorylation sites identified in the three experiments (2). The blue square indicates the total number of proteins and phosphorylation sites for which H/L ratios could be determined rigorously and for which the precise position of the phosphate could be assigned with 95% confidence. The orange square indicates proteins and phosphopeptides containing a minimal consensus motif, and the red square indicates proteins and phosphopeptides that decreased in abundance over 50% after Cdk1 inhibition (log2 H/L < −1). Cdk1 substrates (listed in Table S1) were defined by the overlap between the orange and red squares. Squares are scaled to the number of proteins.