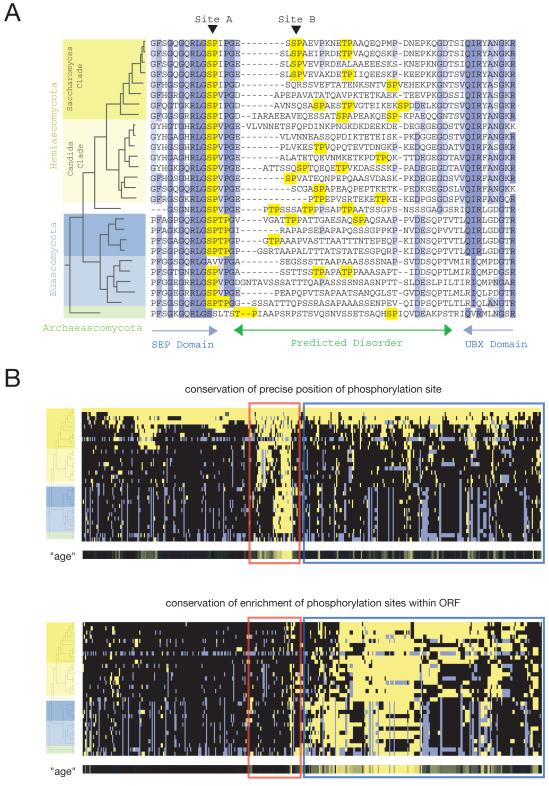
Fig. 4.
Evolution of Cdk1-dependent phosphorylation sites. (A) Representative multiple sequence alignment of 27 orthologs of S. cerevisiae Shp1. The ascomycete phylogeny (Fig. S3) is shown to the left of the alignment. Amino acid conservation is indicated by blue boxes, and minimal Cdk1 consensus motifs are highlighted in yellow. Blue arrows indicate predicted domains and the green arrow indicates a predicted disordered region (PONDR). (B) Hierarchical clusters summarize the evolution of all 547 Cdk1 phosphorylation sites: each row is a different species and each column is a different phosphorylation site. The phylogeny (32 species; see Fig. S3) is represented by a tree at the left. In the top clustergram, yellow indicates that a consensus site (S/T-P) aligns with the phosphorylation site detected in S. cerevisiae (top row). Gray indicates that no single ortholog was detected in that species. In the bottom clustergram, yellow indicates that there is an enrichment of Cdk1 consensus sites in the ortholog of the S. cerevisiae protein we identified as a Cdk1 substrate. Enrichment in each ortholog was assessed by assuming that the expected frequency of a consensus motif is equal to the global frequency across all ORFs in the species, and then using the Poisson distribution to calculate the probability of observing greater than or equal to the actual number of consensus sites. Enrichment was defined by a p-value of less than 0.01 (for example, a typical 400-residue protein is expected to contain 2.8 sites, but must contain 8 or more sites to achieve p<0.01; see Table S5 for details). Two groups are highlighted within the clustergrams: one with conservation of precise site position (red box) and one with conservation of enrichment of consensus sites (blue box). Beneath each clustergram is a metric termed “age”, which summarizes each column as a single conservation score (Fig. S4). More intense yellow indicates greater conservation.