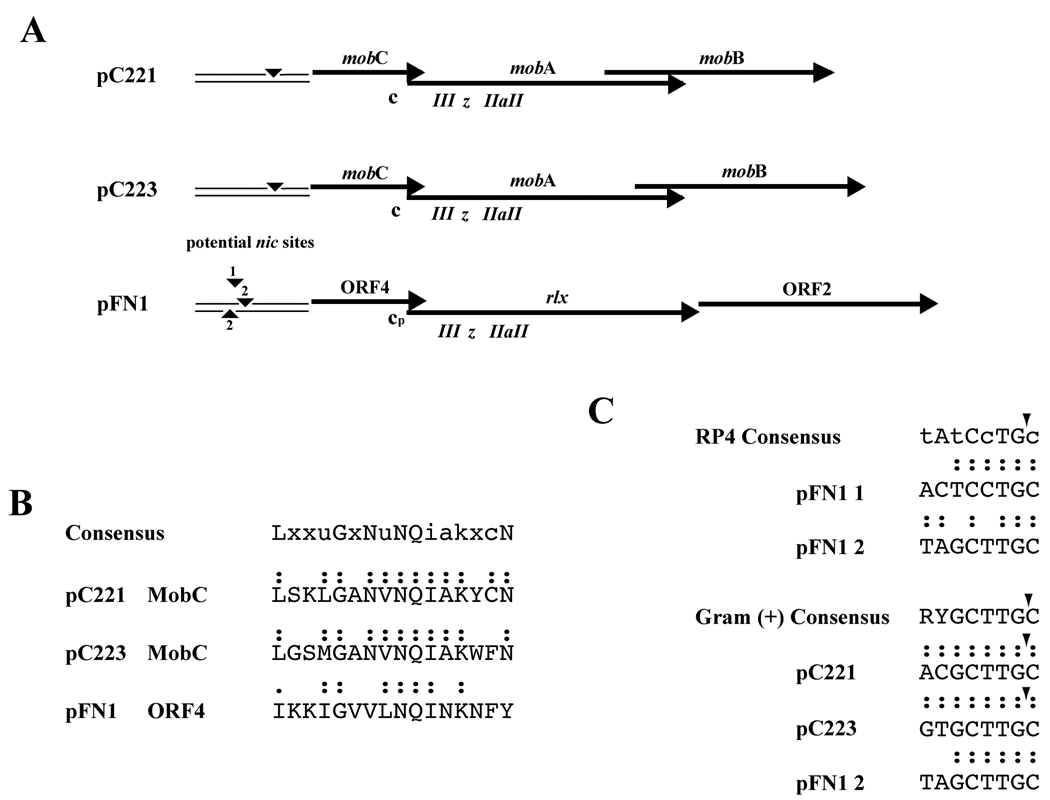
Figure 2. Comparison of pFN1 Mobilon with Staphylococcal Plasmid Mobilons.
(A) Alignment of pFN1 genes and elements predicted to be involved in plasmid mobilization with those of the mobilizable S. aureus plasmid pC221 and pC223. Elements indicated include nic sites (▾) and potential pFN1 nic sites 1 and 2; c, MobC motif; cp, putative pFN1 MobC motif; relaxase motifs, III, z, IIa and II. (B) Alignment of MobC protein motif of pC221 and pC223 (Varsaki et al., 2009) with predicted MobC motif of pFN1 ORF2. Colon (:) indicates identity with consensus; period (.) indicates conservative substitution. (C) Alignment of potential pFN1 nic sites with previously defined consensus sequences for MOBp family nic sites. The RP4 consensus sequence, representative of a Gram-negative nic site, indicates conserved nucleotides in capital letters and semi-conserved nucleotides in lower case letters (Lawley et al., 2004). The Gram-positive consensus is based on an analysis of staphylococcal plasmids (Smith and Thomas, 2004) and for pC221 and pC223 the nic site corresponding to that of the consensus has been demonstrated. The positions of confirmed nic sites (▾) are indicated. Colon (:) indicates identity with consensus.