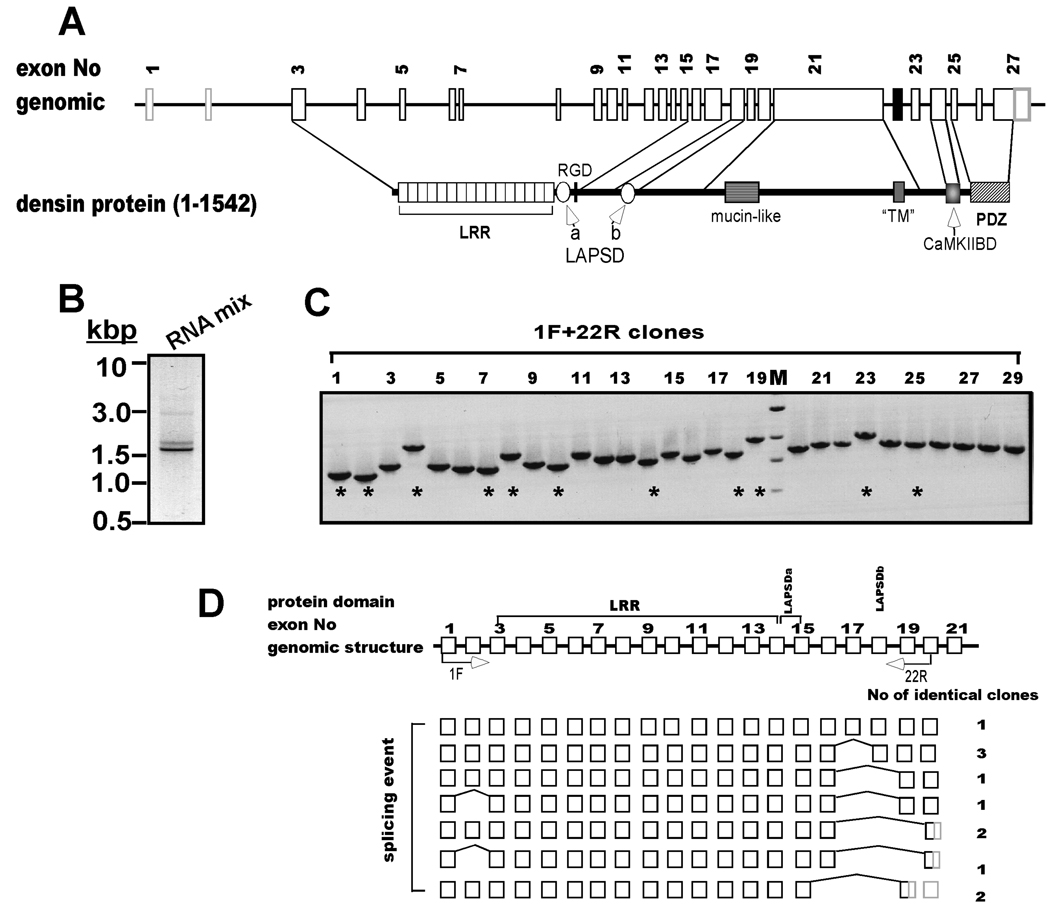
Figure 1. Alternative splicing is detected at the 5’-end of densin.
A. Schematic domain diagram of densin. Structure of densin gene on rat chromosome 2 (GI 34861224) and conserved domains in the full-length form of the protein (1542 amino acids). LRR, leucine-rich repeat; LAPSDa/b, LAP protein specific domains a and b, respectively; RGD: putative integrin interacting motif; “TM”, putative transmembrane domain (see text for discussion); CaMKIIBD, CaMKII binding domain; PDZ, PSD95, dlg1 and ZO1 domain. Sizes of introns, exons and protein domains are depicted to approximate scale. B. Amplification of the 5’ end of the densin mRNA. Full-length cDNA pool was made from a mixture of total mRNAs isolated at different developmental stages by RACE cDNA amplification using primers 1F and 22R (see Experimental Procedures). An aliquot of the product mixture was analyzed on an ethidium bromide-stained gel: an inverted gray-scale image is shown. C. Characterization of individual densin splice variant structures. PCR products from B were inserted into pGEMT Easy vector and 29 isolated bacterial clones containing densin inserts were screened by PCR using 1F and 22R primer set. Products were analyzed on agarose gels stained with ethidium bromide. M: DNA marker: 3.0, 2.0, 1.5 and 1 kbp. D. Summary of densin 5’-splice variant structures. Top: Schematic of the first 21 exons and introns of the densin gene (white boxes and black lines, respectively), not drawn to scale. Labeled arrows below the domain diagram indicate approximate positions of sense and antisense primers used for PCR amplification. DNA sequences of clones (indicated by * in panel C) were aligned to genomic sequence, revealing skipping of 1 or more entire exons (bridging lines). Sequences converted to 3’ untranslated regions (3’ UTRs) by premature stop codons are shaded gray. The number of clones recovered with identical sequences/structures is listed after each variant.