Abstract
Toll-like receptors play important roles in regulating immunity against microbial infections. Toll-like receptor 8 (TLR8) belongs to a subfamily comprising TLR7, TLR8 and TLR9. Human TLR8 mediates anti-viral immunity by recognizing ssRNA viruses, and triggers potent anti-viral and antitumor immune responses upon ligation by synthetic small molecular weight ligands. Interestingly, distinct from human TLR8, mouse TLR8 was not responsive to ligand stimulation in the absence of polyT-oligodeoxynucleotides (polyT-ODN). The molecular basis for this distinct ligand recognition is still unclear. In the present study, we compared activation of TLR8 from different species including mouse, rat, human, bovine, porcine, horse, sheep, and cat by ligand ligations. Only the TLR8s from the rodent species (i.e., mouse and rat TLR8s) failed to respond to ligand stimulation in the absence of polyT-ODN. Multiple sequence alignment analysis suggested that these two rodent TLR8s lack a five-amino-acid motif that is conserved in the non-rodent species with varied sequence. This small motif is located in an undefined region of the hTLR8 ectodomain, immediately following LRR-14. Deletion mutation analysis suggested that this motif is essential for the species-specific ligand recognition of hTLR8, whereas it is not required for self-dimerization and intracellular localization of this receptor.
Keywords: Toll-like receptor 8, Species-specific recognition, Immunotherapy, NF-κB activation, R848, CL075, CL097
1. Introduction
Toll-like receptors (TLRs) are key sensors in innate immune cells for detection of diverse pathogen-associated molecular patterns (PAMPs) from microbes. Activation of TLRs triggers innate immune responses leading to activation of adaptive immune responses for host defense (Beutler et al., 2006; Kumar et al., 2009). To date, ten TLRs (TLR1 to TLR10) have been identified in human cells (Chuang and Ulevitch, 2001). These TLRs are capable of recognizing a variety of structurally diverse microbial pathogens and synthetic ligands, which can be lipids, lipoproteins, glycans, proteins or nucleic acids varying in size from small molecules to macromolecules. In addition, there are species-specific differences in ligand recognition by TLRs from different species. (Beutler et al., 2006; Kumar et al., 2009; Werling et al., 2009).
TLRs are type I transmembrane receptors containing a cytoplasmic Toll/IL-1 receptor (TIR) domain, a transmembrane region, and an ectodomain. The cytoplasmic TIR domain provides a key site for homotypic interaction with TIR domain-containing adapter proteins in the MyD88 family to initiate two major signaling pathways downstream to TLRs: a NF-κB-mediated pathway leading to production of pro-inflammatory cytokines and an IRF-mediated pathway leading to production of IFNs (Carpenter and O’Neill, 2009; Colonna, 2007; Lee and Kim, 2007; West et al., 2006;). The ectodomains of TLRs are characterized by 19 to 25 copies of leucine-rich repeats (LRRs), each consisting of 24 amino acid residues. Based on the three dimensional structure of the TLR3 ectodomain, the first 10 residues of the LRR form a β-sheet and the following 14 residues form a α-helix. These LRRs are arranged to form a horseshoe-shaped solenoid structure with the α-helixes on its outer convex surface, and the parallel β-sheets on its concave surface. The latter provides sites for TLR ligand binding. To date, the molecular determinants in each TLR for the specificity of its ligand binding have not been well characterized (Bell et al., 2003; Jin and Lee, 2008; Werling et al., 2009).
TLR8 belongs to a subfamily comprising TLR7, TLR8 and TLR9. These three TLRs have a higher molecular weight than the other TLRs because of a longer ectodomain that contains 25 copies of LRRs, several large insertions in individual LRRs, and a unique undefined region. TLR7 and TLR8 are phylogenetically closest to each other among all the TLRs, and have high sequence homology (Bell et al., 2003; Chuang and Ulevitch, 2000). As a result, the two receptors overlap in their ligand recognition. They recognize various single-stranded RNAs (ssRNA) derived from viruses, synthetic guanosine- or uridine-rich ssRNA, and synthetic small molecules with a structure related to nucleic acids. Whereas imiquimod, loxoribine, isatoribine, and SM 360320 selectively activate TLR7, others such as CL097, CL075, and R848 activate both receptors. Because their potent immunoadjuvant effects potentially facilitate eradication of virus-infected cells and cancer cells from the body, TLR7/8 agonists are being investigated as anti-viral and antitumor therapies (Agrawal and Kandimalla, 2007; Parkinson, 2008).
Generally, therapeutic agents are tested preclinically in rodent animal models and the resulting data are then transferred to other species for further investigation. However this general strategy may not be applicable to TLR therapeutics, given the species specificity of TLR responses. For example, ligand stimulation of hTLR8 elicits potent immune responses, while mTLR8 is not responsive in the absence of polyT-ODN (Gorden et al., 2006; Hemmi et al., 2002; Jurk et al., 2002). The molecular basis for this distinct activity is not clear. In the present study, we show that a five-amino-acid-residue motif that is conserved with varying sequence in the several non-rodent species including human, is absent in mTLR8 and rat (r)TLR8 that were not responsive to ligand stimulation in the absence of polyT-ODN. The requirement of this motif for hTLR8 properties, including the species-specific ligand recognition, self-dimerization, and intracellular localization was further characterized.
2. Materials and methods
2.1. Reagents and Antibodies
PolyT-ODN (5′-TTTTTTTTTTTTT-3′) was purchased from Sigma (St. Louis, MO). R848 was purchased from GLS Synthesis (Worcester, MA). CL075 and CL097 were purchased from Invivogen (San Diego, CA). Human, mouse, and rat total RNAs were purchased from Clontech (Mountain View, CA). Animals, including bovine, porcine, horse, sheep and cat genomic DNAs were purchased from Zyagen Laboratories (San Diego, CA). Antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
2.2. Cloning of rTLR8
Genomic DNA sequence containing rTLR8 (NW_048039) was aligned with nucleotide sequence of mTLR8 to determine the 5′-end and 3′-end nucleotide sequences of rTLR8. Based on these sequences, primers were designed to clone rTLR8 cDNA by PCR amplification from rat first strand cDNA libraries. This library was prepared from total RNA samples using a SuperScript™ preamplification kit (Invitrogen, Carlsbad, CA). The cDNA sequence was submitted to the GenBank database under the accession number EF032638.
2.3. Expression constructs for TLR8 from different species
Human, mouse and rat TLR8 cDNAs were PCR amplified from their first strand cDNA libraries. Taking advantage of the fact that TLR8 is coded by a single exon except for the first methioine (Astakhova et al., 2009; Chuang and Ulevitch, 2000), DNA fragments containing the complete coding region for bovine, porcine, horse, sheep and cat TLR8 were PCR amplified from their genomic DNA. Gene specific primers for PCR amplification were designed based on the cDNA sequence for complete coding region of TLR8 for human (NM_138636), mouse (NM_133212), rat (EF032638), bovine (EF583902), porcine (NM_214187), horse (NM_001111301), sheep (AM981306), and cat (EF484949).
2.4. Expression constructs for TLR8 mutants and chimeras
DNA fragments containing various mutated and chimeric TLR8 were generated by two-step PCR amplification procedures. To generate point or deletion mutants of hTLR8, a primer containing the desired mutation was used to generate a first DNA fragment. This DNA fragment was then used as a primer to generate a DNA fragment containing the complete coding region of a mutant hTLR8. To generate hTLR8 and mTLR8 chimeras, DNA primers that contain both human and mouse TLR8 sequences at the swapping sites as indicated were designed. DNA fragments containing the hTLR8 part of the chimera were generated in the first PCR amplification using hTLR8 cDNA as template. These PCR products then served as a primer in the second PCR amplification with a second primer designed based on mTLR8 nucleotide sequences and mTLR8 as template to generate DNA fragments containing the hTLR8 and mTLR8 chimeras. These amplified DNA fragments were restriction digested and subcloned into a PEF6 vector to generate expression constructs.
2.5. Cell Culture, transfection, and TLR8 activation reporter assay
HEK 293 cells were maintained in DMEM supplemented with 10% fetal bovine serum. The cells were co-transfected on the following day using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) with TLR8 expression vector, β-galactosidase plasmid, and a NF-κB driven ELAM luciferase-reporter plasmid. Fourteen hours later, the cells were treated with various TLR8 agonists as indicated for 6 h. The cells were lysed, and luciferase activity was determined using reagents from Promega Corp. (Madison, WI). Relative luciferase activities were calculated as fold induction compared to an unstimulated vector control.
2.6. Bioinformatics Analysis
Multiple sequences alignment and phylogenetic analysis for TLR8 from multiple species were performed with a ClustalW2 computer program (http://www.ebi.ac.uk/Tools/clustalw2/; Thompson et al., 1994) and MUSCLE computer program (http://www.ebi.ac.uk/Tools/muscle/; Edgar, 2004).
2.7. Immunoblotting
Cell lysates were resolved on polyacrylamide gels and transferred to PVDF membranes (Immobilon-P, Millpore, Bedford, MA). The membranes were immunoblotted with anti-Flag M2 mAb (Sigma, MO), followed by a HRP-conjugated secondary antibody (Cell Signaling, Danvers, MA). The immunoreactive bands were detected with ECL+Plus western blotting reagents (Amersham Pharmacia Biotech, Piscataway, NJ).
2.8. Immunofluorescence staining
Cells were fixed in 2% paraformaldehyde and permeabilized with 0.2% of Triton X-100 in PBS. TLR8 was visualized by staining with an anti-Flag M2 monoclonal antibody, followed by an Alexa 488-labeled anti-mouse antibody (Invivogen, Carlsbad, CA). Cell nuclei were visualized using 5 ng/ml DAPI (Invivogen, Carlsbad, CA). Samples were observed with an IX70 inverted immunofluorescence microscope (40x) (Olympus America, Melville, NY).
2.9. Statistical Analysis
Groups of data are expressed as mean±SD. Statistical analyses were performed using Student’s t-test. All groups were from three or more independent experiments. p< 0.05 was considered statistically significant.
3. Results
3.1. Mouse and Rat TLR8 display a similar ligand response
To investigate whether other rodent TLR8s display a similar ligand response to that of mice, we cloned rTLR8 cDNA and compared its protein sequence and ligand response with hTLR8 and mTLR8. Human, mouse and rat TLR8 contain 1041, 1032 and 1029 amino acid residues, respectively (Fig. 1). The phylogenetic analysis showed that among the TLR8 sequences analyzed, mouse and rat TLR8 are most closely related to each other. These two TLR8s share a 94.0% protein sequence similarity, compared to the 82.2%, 80.7%, 81.0% 80.7%, 82.9% and 82.0% similarity between the mouse and human, porcine, bovine, sheep, horse and cat TLR8, respectively (Supplementary data, Fig. S1). To assess the ligand response, cells co-transfected with human, mouse or rat TLR8 and an ELAM luciferase-reporter gene (Schindler and Baichwal, 1994) were treated with CL075 or polyT-ODN alone, or in combination, and then tested for NF-κB-dependent luciferase activity. Similar to the mTLR8, rTLR8 could not be activated by CL075 or polyT-ODN alone but was activated by the combination of CL075 and poly13T-ODN. These results showed that both mouse and rat TLR8 exhibit a similar ligand response (Fig. 2).
Fig. 1.
Alignment of human, mouse and rat TLR8 proteins. Amino acids were color coded to indicate their chemical properties: blue indicates acidic; green indicates hydroxyl/amine/basic/Q; pink indicates basic; red indicates hydrophobic (including aliphatic Y). Identical residues were represented by an asterisk, conservative substitutions by a single dot, and highly conservative substitutions by two dots. The ectodomain, transmembrane domain and cytoplasmic domain in hTLR8 span amino acid residues 1–828, 829–852, and 853–1041, respectively. The LRRs are underlined, and the undefined region is double underlined. The PGIQ and RQSYA motifs are boxed.
Fig. 2.
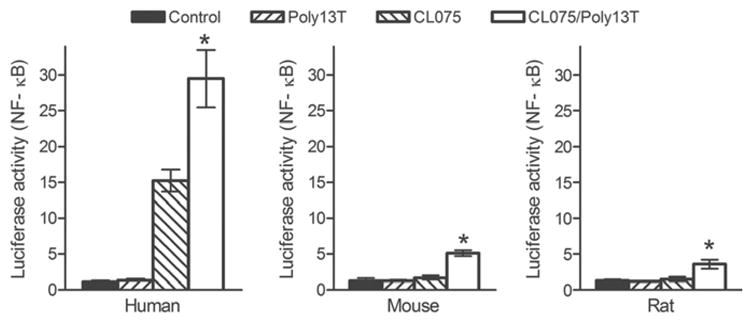
Activation of human, mouse and rat TLR8 by ligand in the absence and presence of polyT-ODN. HEK293 cells were transfected with expression vector for TLR8 from human, mouse or rat as indicated plus an ELAM luciferase-reporter gene, and treated with 2μM of CL075 or 10μM of poly13T-ODN either separately or in combination. Relative luciferase activities were determined. Data shown represent mean ± SD (n=3). *P < 0.05 vs. control cells.
3.2. Distinct ligand response between non-rodent and rodent TLR8
To further explore the species specificity of the TLR8 response, we compared the responses between the two rodent TLR8s and TLR8s from human, porcine, bovine, sheep, horse and cat to agonists including CL075, CL095 and R848. As shown in Fig. 3, these TLR8 ligands induced NF-κB activities to differing degrees in cells transfected with human, porcine, bovine, sheep, horse or cat TLR8 expression vector, but not in cells transfected with mTLR8 or rTLR8. The lack of mTLR8 and rTLR8 activation was not dependent on the dose of the ligand used, since increasing the concentration of CL075, CL097 and R848 up to 9 μM did not enhance their responsiveness to ligand stimulation (Supplementary data, Fig. S2). Thus, there is a distinct TLR8 response between the non-rodent and rodent species, with neither mTLR8 nor rTLR8 responding to ligand stimulation in the absence of polyT-ODN.
Fig. 3.
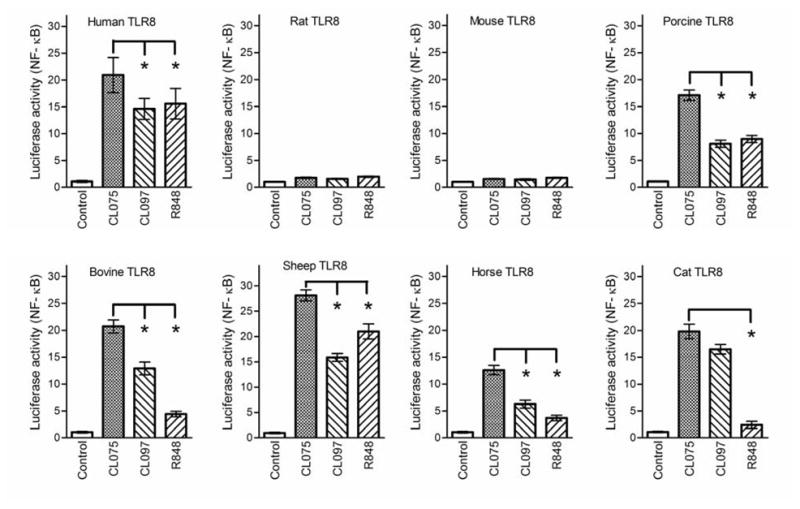
Activation of TLR8 from non-rodent and rodent species. HEK293 cells were transfected with expression vector for TLR8 from different species as indicated plus an ELAM luciferase-reporter gene, and treated with 2μM of CL075, CL097 and R848. Relative luciferase activities were determined. Data shown represent mean ± SD (n=3). *P < 0.05 vs. CL075 stimulated cells.
3.3. Ectodomain determines the species-specific response of TLR8
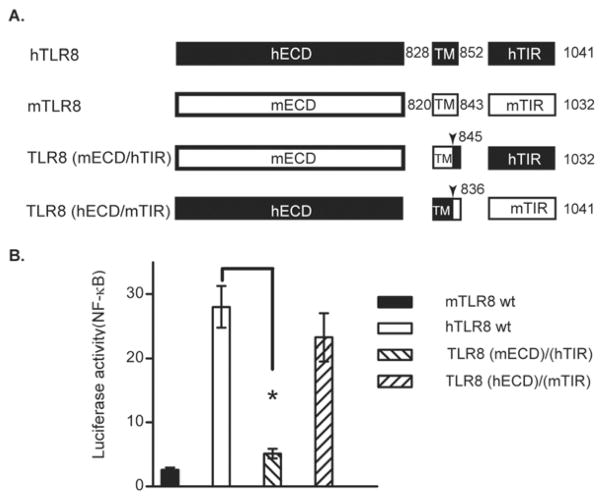
To investigate the molecular determinant underlying this distinct response, we swapped the ectodomain (ECD) and cytoplasmic domain (TIR) between the human and mouse TLR8 to generate a TLR8(mECD/hTIR) and a TLR8(hECD/mTIR) chimera, and tested their activation by ligand ligation. The results indicated that cells transfected with hTLR8 and the TLR8(hECD/mTIR) chimera generated a comparable response to CL075 stimulation, whereas the mTLR8 and the TLR8(mEDC/hTIR) chimera transfected cells failed to respond (Fig. 4). These results suggested that while the cytoplasmic domains of human and mouse TLR8 have a comparable ability to initiate downstream signaling cascades, their ectodomains determine whether signaling is initiated in response to ligand stimulation. Thus, the molecular determinant for species-specific response between human and mouse TLR8 is located within their ectodomains.
Fig. 4.
Role of the ectodomain in mediating TLR8 activation. (A) A schematic of the human and mouse TLR8 chimera constructs. ECD, extracellular domain/ectodomain; TM, transmembrane domain; TIR, Toll-interleukin 1 receptor domain. (B) Ectodomain determines response of TLR8 to ligand stimulation. HEK293 cells were transfected with different TLR8 expression vectors as indicated, plus ELAM luciferase-reporter gene, and stimulated with 2 μM of CL075. Relative luciferase activities were determined. Data shown represent mean ± SD (n=3). *P < 0.05 vs. cells transfected with hTLR8 wt plasmid.
3.4. Molecular determinant in ectodomain for hTLR8 activation
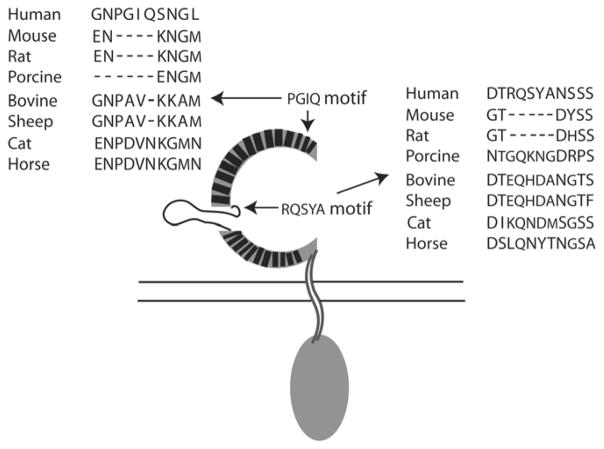
The results from cell-based TLR8 activation assay suggested that both rodent TLR8s may lack some common molecular determinant in their ectodomain that is required for the distinct ligand recognition by non-rodent TLR8s. Alignment of the hTLR8, mTLR8 and rTLR8 sequences showed that these proteins have high homology through the whole protein sequence, except for a PGIQ motif and a RQSYA motif in hTLR8 that were missing from the ectodomains of the mouse and rat TLR8s (Fig. 1). The PGIQ motif is located in LRR-2 of the 25 LRRs found in the ectodomain of hTLR8, and the RQSYA motif is located in the undefined region immediately following LRR-14. We expanded this alignment analysis by including additional TLR8 protein sequences from non-rodent species including porcine, bovine, sheep, cat and horse. The results indicated that the PGIQ motif was also absent in porcine TLR8, but was conserved in human, horse and cat TLR8 and partially conserved in bovine and sheep TLR8 although the amino acid sequence varied between species (Fig. 5). Interestingly, the RQSYA motif identified in the undefined region of hTLR8 is missing only in the mouse and rat TLR8 but conserved with varying amino acid sequences in the non-rodent TLR8s, consistent with the different ligand response capabilities of the two groups of TLR8 (Fig. 3 and Fig. 5). This suggests that this five-amino-acid motif may be involved in the species-specific ligand recognition of the non-rodent TLR8s.
Fig. 5.
Location and alignment of the PGIQ and RQSYA motifs in TLR8s. The PGIQ motif is located in LRR-2. The RQSYA motif is located in the undefined region immediately following LRR-14. Sequence alignments show the corresponding regions of these motifs in TLR8 from different species. In the schematic of hTLR8, dark boxes represent the LRRs and the loop structure represents the undefined region.
3.5. The RQSYA motif in an undefined region is required for hTLR8 activation
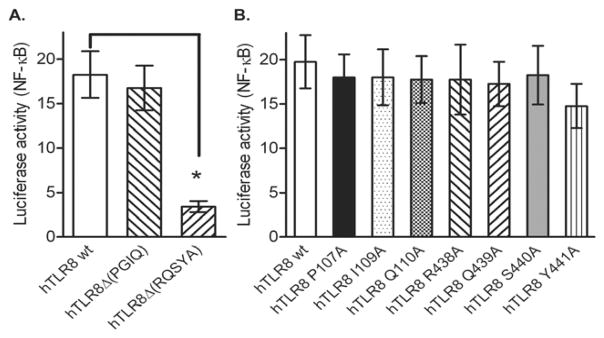
To investigate the requirement for these two motifs in hTLR8 activation, we generated deletion mutants of hTLR8 that lack each of the motifs. Deletion of the RQSYA motif from hTLR8 abrogated its response to ligand stimulation, whereas deletion of the PGIQ motif had no effect on activation (Fig. 6A). A glutamine residue is conserved in the five-amino-acid motif in the non-rodent TLR8s analyzed (Fig. 5). We further generated point mutants for this glutamine and the other amino acid residues within these two motifs of hTLR8 to determine whether any of them is required for hTLR8 activation. The results indicated that neither the glutamine nor any other residue in this motif were essential for hTLR8 activation (Fig. 6B). These results suggested that the RQSYA motif is required for hTLR8 activation, although individual residues in this motif may not be involved in ligand interaction.
Fig. 6.
Requirement for the RQSYA motif for TLR8 activation. HEK293 cells were transfected with expression vector for wild type, and (A) deletion mutated hTLR8, or (B) point mutated hTLR8, plus ELAM luciferase-reporter gene. The cells were stimulated with 2 μM of CL075 and relative luciferase activities were determined. Data shown represent mean ± SD (n=3). *P < 0.05 vs. cells transfected with hTLR8 wt plasmid.
3.6. The RQSYA motif is not required for hTLR8 dimerization and intracellular localization
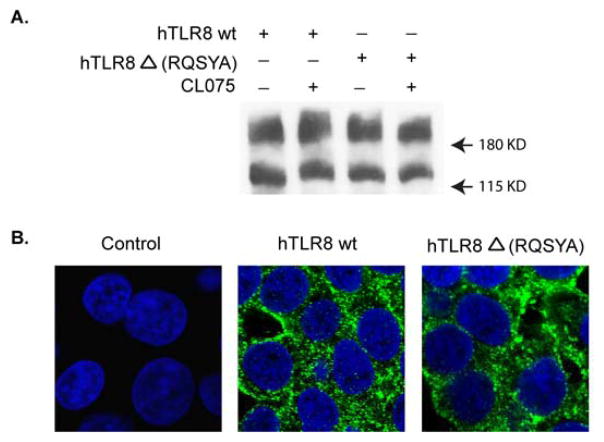
In addition to its ability to respond to ligand stimulation, other properties of TLR8 include its self-dimerization and intracellular localization. Self-dimerization occurs in TLRs and can be detected even under denaturing conditions. Ligand-induced dimerization was suggested as a mechanism for TLR2 and TLR4 activation, whereas bTLR8 and hTLR9 were shown to form dimers prior or after ligand stimulation (Jin et al., 2007; Kim et al., 2007; Latz et al., 2007; Zhu et al., 2009). Distinct from cell surface expression of the other TLRs, TLR3, TLR7, TLR8 and TLR9 are localized in intracellular vesicles including the endoplasmic reticulum, endosomes, and lysosomes (Beutler et al., 2006; Kumar et al., 2009). We investigated the effect of deleting the RQSYA motif on these properties of hTLR8 by expressing the RQSYA deletion mutant and wild type hTLR8 in cells. Expression of hTLR8 and self-dimerized TLR8 was detected by immunoblot at around 160 kDa and 250 kDa, respectively. Cellular localization of these TLR8s was determined by immunofluorescence staining. The results indicated that deletion of the RQSYA motif had no effect on the ability of hTLR8 to form a dimer in the presence or absence of ligand stimulation (Fig. 7A), or on its intracellular localization (Fig. 7B) suggesting that, in contrast to its effect on hTLR8 activation, this motif is not essential for these two properties of this receptor.
Fig. 7.
Deletion of the RQSYA motif has no effect on TLR8 dimerization and intracellular localization. HEK293 cells were transfected with expression vector for hTLR8 or hTLR8Δ(RQSYA). (A) hTLR8 dimerization was analyzed by immunoblot analysis using anti-Flag M2 mAb. (B) Cellular localization of TLR8 was visualized by immunofluorescence staining using anti-Flag M2 mAb, followed by an Alexa 488-labeled anti-mouse antibody (green). Nuclei were stained by DAPI (blue).
4. Discussion
Species-specific ligand responses are frequently seen between TLRs from different species. For example, both triacyl-lipopeptides and diacyl-lipopeptides activate a bovine(bo) TLR2 and boTLR1 complex, in contrast to the hTLR2/1 and hTLR2/6 receptor complexes activated respectively in human cells. Rhodobacter sphaeoides activates TLR4 signaling in horse and hamster, but acts as an antagonist in human and mouse. Chicken(ch) TLR5 generates stronger responses to flagellin from S. enterica serovar Typhimurium than hTLR5. Activation of hTLR8 elicites potent immune responses, while mTLR8 is not responsive to ligand stimulation in the absence of polyT-ODN. CpG-ODN containing a GTCGTT motif preferentially activates hTLR9, whereas a GACGTT motif displays the greatest activity toward mouse(m) TLR9 (Werling et al., 2009). However, the molecular basis underlying the species-specific recognition are rarely investigated. In the present study, we investigated the molecular determinant governing the species-specific ligand recognition of TLR8.
We analyzed the response of TLR8 from eight species (mouse, rat, human, porcine, bovine, sheep, horse, and cat). Non-rodent TLR8s respond to ligand stimulation with differing magnitudes of activation. In contrast, the two rodent TLR8s, mouse and rat TLR8 that share 94% similarity, fail to respond to ligand stimulation in the absence of polyT-ODN. This is consistent with previous reports that TLR8-specific RNA sequences are not able to activate cytokine production in immune cells isolated from these two species except in the presence of polyT-ODN (Forsbach et al., 2008; Hemmi et al., 2002; Jurk et al., 2002). Two possible explanations for the non-responsiveness of rodent TLR8s are that (1) both non-rodent and rodent TLR8s bind ligands with equal affinity but rodent TLR8s are weaker signaling initiators compared to non-rodent TLR8s, and (2) whereas TLR8s have equal capability to initiate intracellular signaling, there is distinct ligand recognition between TLR8s from different species. Using a chimera strategy, we showed that the cytoplasmic domain of human and mouse TLR8 have comparable activity in initiating downstream signaling upon ligand stimulation. Instead, the distinct ligand responses between hTLR8 and mTLR8 suggested that the molecular determinant governing this distinct ligand recognition is located within the ectodomain. We identified a RQSYA motif that is conserved with varying amino acid sequences in the ectodomain of non-rodent TLR8s, but was absent in rodent TLR8s. This five-amino-acid motif is required for the ligand recognition of hTLR8, whereas it is not essential for self-dimerization and intracellular localization. These results suggested that this five-amino-acid motif could be the molecular determinant for the distinct ligand recognition between non-rodent and rodent TLR8s.
Architectural analysis of the ectodomain of hTLR8 revealed 25 copies of LRRs, large insertions in several individual LRR units, and an undefined region that is inserted between LRR-14 and LRR-15 (Bell et al., 2003). The insertions in LRR-2,-5,-8, -11, and -20 are essential for activation of boTLR8 by ligands (Zhu et al., 2009), and certain amino acid residues in the LRR-8 and LRR-17 are required for hTLR8 activation (Gibbard et al., 2006). However, the role in TLR activation of the undefined region, which is unique to the TLR7/8/9 subfamily, has not been explored. The RQSYA motif that we identified is located in the undefined region in hTLR8 immediately following LLR-14. Deletion of this motif abrogated hTLR8 responses to ligand stimulation, suggesting an essential role of this undefined region in TLR8 activation. However, mutation of each individual amino acid residue in this motif to alanine had no effect on hTLR8 activation, suggesting that these residues may not be involved in a direct interaction with ligands. This is consistent with the finding that the sequence of this motif varies in different non-rodent TLR8 without affecting activation of these TLR8s (Fig. 2 and Fig. 5). Since this motif is located at the junction region of LLR-14 and the undefined region, which is at the deepest region of the solenoid concave, a reasonable speculation is that this motif may play a role as a hinge for building a three dimensional conformation for a high affinity binding to ligands.
In addition to the species-specific ligand recognition between the two groups of TLR8 from non-rodent and rodent species, different ligand recognition patterns were observed for each of the non-rodent TLR8s. For example, while CL075 and CL097 induced 15- to 20-fold activation of the human, bovine, sheep, and cat TLR8, the responses to R848 were more varied. R848 induced human and sheep TLR8 10- to 15-fold, whereas bovine and cat TLR8 were induced less than 5-fold. These multiple layers of species-specific differences among the TLR8s suggest the existence of multiple molecular determinants for ligand binding in the ectodomain of TLR8. Again, this is consistent with the results from previous studies (Gibbard et al., 2006; Zhu et al., 2009) and the present study that the insertion regions within individual LRRs and the undefined region are essential for TLR8 activation.
Through evolution, TLRs are under strong selection to maintain inherent functions and to adopt new functions. The transmembrane domain and the cytoplasmic TIR domain that initiate intracellular signaling for host defense responses are highly conserved among different species and different TLRs. By contrast, the ectodomains of TLRs exhibit significantly higher divergence, reflecting their involvement in recognition of diversified molecular patterns of ligands from multiple microbial sources (Roach et al., 2005; Zhou et al., 2007; Temperley et al., 2008). Our findings, that initiation of downstream signaling by the cytoplasmic domains of both human and mouse TLR8 is conserved, and that the RQSYA motif essential for the species-specific activity of hTLR8 is located in the ectodomain are consistent with the rules for guiding the evolution of TLRs. The absence of this motif for the species-specific recognition in rodent TLR8s suggests that it may have been deleted in the mouse and rat TLR8 during a species-specific co-evolution.
In addition to the mouse and rat TLR8, other TLR8s that are unable to respond to ligand stimulation include rabbit, chicken, and duck TLR8 that are reported as pseudogenes either because their coding region is disrupted, or because no transcripts are detectable (Astakhova et al., 2009; Guo et al., 2008; Philbin et al., 2005). TLR8 has high protein homology with TLR7 and these two TLRs have overlapping ligand recognition and functions in induction of immune responses (Agrawal and Kandimalla, 2007; Parkinson, 2008). In most mammals, both genes are located on the X chromosome, separated by a distance of less than 100 kb between the coding exons of the two genes (Astakhova et al., 2009; Chuang and Ulevitch, 2000), raising speculation as to whether TLR8 is a redundant gene through evolution.
Overall, the results presented in this study show the diversified ligand recognition pattern among TLR8s from different species, and provide a molecular basis to explain why the two rodent TLR8s display low activities in response to ligand stimulation.
Supplementary Material
Acknowledgments
This work was support in part by grants from the National Institutes of Health GM069652 (T.H.C.), California Tobacco-Related Disease Research Program 16RT-0103 (T.H.C.), National Science Council NSC-96-2321-B-002-034-MY2 (L.C.H.), and National Natural Science Foundation of China 30572116 and 30672389 (R.X.)
Abbreviations
- TLR
Toll-like receptor
- NF-κB
nuclear factor-κB
- LRR
leucine rich repeat
- ECD
ectodomain
- TIR
Toll-interleukin 1 receptor domain
- ELAM
endothelial-leukocyte adhesion molecule
- HEK
human embryonic kidney
- DAPI
4′, 6 diamidino-2-2 phenylindole
- polyT-ODN
polyT-oligodeoxynucleotides
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agrawal S, Kandimalla ER. Synthetic agonists of Toll-like receptors 7, 8 and 9. Biochem Soc Trans. 2007;35:1461–1467. doi: 10.1042/BST0351461. [DOI] [PubMed] [Google Scholar]
- Astakhova NM, Perelygin AA, Zharkikh AA, Lear TL, Coleman SJ, MacLeod JN, Brinton MA. Characterization of equine and other vertebrate TLR3, TLR7, and TLR8 genes. Immunogenetics. 2009;61:529–539. doi: 10.1007/s00251-009-0381-z. [DOI] [PubMed] [Google Scholar]
- Bell JK, Mullen GE, Leifer CA, Mazzoni A, Davies DR, Segal DM. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 2003;24:528–533. doi: 10.1016/s1471-4906(03)00242-4. [DOI] [PubMed] [Google Scholar]
- Beutler B, Jiang Z, Georgel P, Crozat K, Croker B, Rutschmann S, Du X, Hoebe K. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu Rev Immunol. 2006;24:353–389. doi: 10.1146/annurev.immunol.24.021605.090552. [DOI] [PubMed] [Google Scholar]
- Carpenter S, O’Neill LA. Recent insights into the structure of Toll-like receptors and post- translational modifications of their associated signalling proteins. Biochem J. 2009;422:1–10. doi: 10.1042/BJ20090616. [DOI] [PubMed] [Google Scholar]
- Chuang TH, Ulevitch RJ. Cloning and characterization of a sub-family of human toll-like receptors: hTLR7, hTLR8 and hTLR9. Eur Cytokine Netw. 2000;11:372–378. [PubMed] [Google Scholar]
- Chuang TH, Ulevitch RJ. Identification of hTLR10: a novel human Toll-like receptor preferentially expressed in immune cells. Biochim Biophys Acta. 2001;1518:157–161. doi: 10.1016/s0167-4781(00)00289-x. [DOI] [PubMed] [Google Scholar]
- Colonna M. TLR pathways and IFN-regulatory factors: to each its own. Eur J Immunol. 2007;37:306–309. doi: 10.1002/eji.200637009. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsbach A, Nemorin JG, Montino C, Muller C, Samulowitz U, Vicari AP, Jurk M, Mutwiri GK, Krieg AM, Lipford GB, Vollmer J. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J Immunol. 2008;180:3729–3738. doi: 10.4049/jimmunol.180.6.3729. [DOI] [PubMed] [Google Scholar]
- Gibbard RJ, Morley PJ, Gay NJ. Conserved features in the extracellular domain of human toll-like receptor 8 are essential for pH-dependent signaling. J Biol Chem. 2006;281:27503–27511. doi: 10.1074/jbc.M605003200. [DOI] [PubMed] [Google Scholar]
- Gorden KK, Qiu XX, Binsfeld CC, Vasilakos JP, Alkan SS. Cutting edge: activation of murine TLR8 by a combination of imidazoquinoline immune response modifiers and polyT oligodeoxynucleotides. J Immunol. 2006;177:6584–6587. doi: 10.4049/jimmunol.177.10.6584. [DOI] [PubMed] [Google Scholar]
- Guo X, Branton WG, Moon DA, Xia J, Macdonald MR, Magor KE. Dendritic cell inhibitory and activating immunoreceptors (DCIR and DCAR) in duck: Genomic organization and expression. Mol Immunol. 2008;45:3942–3946. doi: 10.1016/j.molimm.2008.06.014. [DOI] [PubMed] [Google Scholar]
- Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, Horiuchi T, Tomizawa H, Takeda K, Akira S. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, Lee H, Lee JO. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Jin MS, Lee JO. Structures of the toll-like receptor family and its ligand complexes. Immunity. 2008;29:182–191. doi: 10.1016/j.immuni.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Jurk M, Heil F, Vollmer J, Schetter C, Krieg AM, Wagner H, Lipford G, Bauer S. Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat Immunol. 2002;3:499. doi: 10.1038/ni0602-499. [DOI] [PubMed] [Google Scholar]
- Kim HM, Park BS, Kim JI, Kim SE, Lee J, Oh SC, Enkhbayar P, Matsushima N, Lee H, Yoo OJ, Lee JO. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Akira S. Pathogen recognition in the innate immune response. Biochem J. 2009;420:1–16. doi: 10.1042/BJ20090272. [DOI] [PubMed] [Google Scholar]
- Latz E, Verma A, Visintin A, Gong M, Sirois CM, Klein DC, Monks BG, McKnight CJ, Lamphier MS, Duprex WP, Espevik T, Golenbock DT. Ligand-induced conformational changes allosterically activate Toll-like receptor 9. Nat Immunol. 2007;8:772–779. doi: 10.1038/ni1479. [DOI] [PubMed] [Google Scholar]
- Lee MS, Kim YJ. Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu Rev Biochem. 2007;76:447–480. doi: 10.1146/annurev.biochem.76.060605.122847. [DOI] [PubMed] [Google Scholar]
- Parkinson T. The future of toll-like receptor therapeutics. Curr Opin Mol Ther. 2008;10:21–31. [PubMed] [Google Scholar]
- Philbin VJ, Iqbal M, Boyd Y, Goodchild MJ, Beal RK, Bumstead N, Young J, Smith AL. Identification and characterization of a functional, alternatively spliced Toll-like receptor 7 (TLR7) and genomic disruption of TLR8 in chickens. Immunology. 2005;114:507–521. doi: 10.1111/j.1365-2567.2005.02125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach JC, Glusman G, Rowen L, Kaur A, Purcell MK, Smith KD, Hood LE, Aderem A. The evolution of vertebrate Toll-like receptors. Proc Natl Acad Sci U S A. 2005;102:9577–9582. doi: 10.1073/pnas.0502272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler U, Baichwal VR. Three NF-kappa B binding sites in the human E-selectin gene required for maximal tumor necrosis factor alpha-induced expression. Mol Cell Biol. 1994;14:5820–5831. doi: 10.1128/mcb.14.9.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temperley ND, Berlin S, Paton IR, Griffin DK, Burt DW. Evolution of the chicken Toll- like receptor gene family: a story of gene gain and gene loss. BMC Genomics. 2008;9:62. doi: 10.1186/1471-2164-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling D, Jann OC, Offord V, Glass EJ, Coffey TJ. Variation matters: TLR structure and species-specific pathogen recognition. Trends Immunol. 2009;30:124–130. doi: 10.1016/j.it.2008.12.001. [DOI] [PubMed] [Google Scholar]
- West AP, Koblansky AA, Ghosh S. Recognition and signaling by toll-like receptors. Annu Rev Cell Dev Biol. 2006;22:409–437. doi: 10.1146/annurev.cellbio.21.122303.115827. [DOI] [PubMed] [Google Scholar]
- Zhou H, Gu J, Lamont SJ, Gu X. Evolutionary analysis for functional divergence of the toll- like receptor gene family and altered functional constraints. J Mol Evol. 2007;65:119–123. doi: 10.1007/s00239-005-0008-4. [DOI] [PubMed] [Google Scholar]
- Zhu J, Brownlie R, Liu Q, Babiuk LA, Potter A, Mutwiri GK. Characterization of bovine Toll-like receptor 8: ligand specificity, signaling essential sites and dimerization. Mol Immunol. 2009;46:978–990. doi: 10.1016/j.molimm.2008.09.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.