Abstract
Three classes of IgG have been described for camelids. IgG1 has a conventional four-chain structure, while IgG2 and IgG3 do not incorporate light chains. The structures and antigen-binding affinities of the so-called heavy-chain classes have been studied in detail; however, their regulation and effector functions are largely undefined. The aim of this study was to examine the participation of conventional and heavy-chain IgG antibodies in the camelid immune defense directed against West Nile virus (WNV). We found that natural infection or vaccination with killed WNV induced IgG1 and IgG3. Vaccination also induced IgG1 and IgG3; IgG2 was produced during the anamnestic response to vaccination. When purified IgGs were tested in plaque-reduction neutralization titer (PRNT) tests, IgG3 demonstrated PRNT activities comparable to those of conventional IgG1. In contrast, IgG2 demonstrated only suboptimal activity at the highest concentrations tested. Flow cytometric analysis revealed that macrophages bound IgG1, IgG2, and IgG3. Furthermore, subneutralizing concentrations of all three isotypes enhanced WNV infection of cultured macrophages. Our results document distinctions in regulation and function between camelid heavy-chain isotypes. The reduced size and distinct structure of IgG3 did not negatively impact its capacity to neutralize virus. In contrast, IgG2 appeared to be less efficient in neutralization. This information advances our understanding of these unusual antibodies in ways that can be applied in the development of effective vaccines for camelids.
The canonical structure of immunoglobulin is a heterodimeric tetramer comprised of heavy and light chains. Camelids produce these conventional antibodies, but they also produce antibodies that do not incorporate light chains (14, 29). Camelid heavy-chain antibodies (HCAbs) comprise 45% of serum IgG in Lama spp. and 75% of serum IgG in Camelus spp. (14). This representation is compatible with a significant role in immune defense; however, functional information concerning these unusual antibodies is scarce.
Antibody function is dictated by structure and binding affinity. Camelid HCAbs are bivalent, with variable domains that undergo somatic hypermutation and exhibit nanomolar binding affinities (8, 10). The H chains lack CH1, the first constant domain that normally associates with the CL domain to stabilize the heterodimer (20, 34). The heterodimer of conventional antibodies is further stabilized by hydrophobic amino acids on the opposing surfaces of the VL and VH domains. In contrast, the VH of HCAbs (VHH) incorporates hydrophilic amino acids at the putative H-L chain interface, conferring solubility on the antigen-binding domain (21). To increase diversification of the combining sites of VHH, the first and second hypervariable loops adopt noncanonical loop structures (8, 9, 11, 26). Furthermore, similar to the case for bovine immunoglobulins, VHH domains may possess an extended complementarity determining region (CDR) 3 that may serve to increase the surface area for antigen binding (33). With masses of approximately 15 kDa, VHH domains are the smallest natural antigen-binding fragments known (14, 19).
The effector function of an immunoglobulin is largely determined by the CH2 and CH3 domains. In camelids, IgG1 is a conventional antibody and IgG2 and IgG3 are HCAbs. Sequence comparisons among llama IgGs indicated that CH2 and CH3 exons are 88 to 98% and 93 to 96% identical, respectively (14, 30). Despite this similarity, functional distinction is evident in that IgG1 and IgG3 bind protein G, while IgG2 does not (6, 14, 30). Since it is the Fc portion of IgGs that is recognized by protein G (2), this differential binding suggests conformational distinctions among the isotypes that may correlate with FcR-dependent effector functions. Furthermore, HCAbs have molecular masses of approximately 100 kDa, compared with 150 kDa for IgG1. The reduced mass, in combination with the small size of the VHH domain, may enable HCAbs to bind epitopes that would be inaccessible to conventional antibodies.
The novel structural features of HCAbs suggest that these isotypes may have distinct roles in camelid immunity. Research into camelid antibody functions has been hindered greatly by the lack of specific reagents. To address this, we produced and characterized mouse monoclonal antibodies (MAbs) that bind specifically to IgG1, IgG2, or IgG3 of llamas and alpacas (6). In this study, we used these reagents to compare the contributions and functions of IgG1, IgG2, and IgG3 in immune responses elicited in alpacas by vaccination, natural infection, and experimental infection with a flavivirus, West Nile virus (WNV).
MATERIALS AND METHODS
Animals and sera.
Sera from alpacas that were naturally infected with WNV (n = 4) were obtained during a surveillance study conducted in New Jersey. Sera from alpacas vaccinated against WNV (n = 8 vaccinated and 4 vaccinated and boosted animals) were collected as part of a vaccine study that has been published previously (16). In all cases, camelid blood was collected by jugular venipucture into Vacutainer tubes containing no anticoagulant. Sera were separated by centrifugation and stored at −20°C.
For macrophage cultures, blood was obtained from two adult female alpacas housed at Cornell University. Animal care was performed in compliance with the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care.
WNV vaccination.
The vaccination protocol has been described elsewhere (16). Briefly, alpacas were vaccinated with 1 ml of formalin-inactivated WNV vaccine (West Nile Innovator; Fort Dodge Animal Health), administered intramuscularly. Alpacas received three vaccinations at 3-week intervals. Some animals received an additional dose of vaccine 1 year after the third immunization and are referred to as “vaccinated/boosted” animals. Sera from these animals were collected 4 weeks after the fourth immunization.
Antibodies and antigens.
Polyclonal goat anti-llama IgG(H+L) conjugated to horseradish peroxidase (HRP) (Bethyl Laboratories Inc.) was used for enzyme-linked immunosorbent assay (ELISA) and to form immune complexes with alpaca IgG. Mouse MAbs were detected with HRP-conjugated goat anti-mouse IgG(H+L) (ICN/Cappel). The anti-IgG1 (27E10 and 28G4), anti-IgG2 (19D8 and 16A4), and anti-IgG3 (8E1 and 2B11) MAbs have been described elsewhere (6).
Recombinant WNV envelope (E) and nonstructural (NS1 and NS5) proteins were prepared by cloning reverse transcription-PCR (RT-PCR)-amplified DNA fragments into pTriEx-1 plasmid vectors (Novagen). Baculovirus recombinants were prepared by cotransfecting plasmid DNA and BacVector 3000 DNA (Novagen) and were used to infect Sf9 cells. Cells were lysed, and expressed proteins were purified using nickel-affinity chromatography (Bio-Rad).
Detection of llama IgGs specific for WNV antigens.
The ELISA described by Daley et al. (6) was modified to detect camelid IgGs specific for WNV. Well volumes were 25 μl, and plates were incubated for 1 h at room temperature unless otherwise specified. Plates were coated overnight at 4°C with 5.0 μg/ml WNV E, NS1, or NS5 protein, diluted in 10% Dulbecco's phosphate-buffered saline (DPBS). Wells were blocked with DPBS containing 2% skim milk and 0.05% Tween 20. Plates were washed three times with DPBS containing 0.05% Tween 20 after each incubation step. All sera and antibodies were diluted in blocking solution. Alpaca sera were diluted 1:100 and tested in triplicate. Plates were washed, and wells were incubated with the 27E10, 19D8, or 8E1 MAb (5.0 μg/ml) or with HRP-conjugated goat anti-llama IgG (0.1 μg/ml) as a positive control. After being washed, wells were incubated with 5.0 μg/ml HRP-conjugated goat anti-mouse IgG diluted in blocking solution containing 10% normal goat serum. The assay was developed with 3,3′,5,5′-tetramethylbenzidine (TMB; KHL), and reactions were terminated by adding 1 M H3PO4. The optical density (OD) was measured at 450 nm with a microplate reader (Biokinetics reader, model EL340; Bio-Tek Instruments).
Purification of camelid IgGs.
IgGs were purified using affinity columns prepared with MAb 27E10, 19D8, or 8E1, as described elsewhere (6). Purified IgG fractions were dialyzed against DPBS and evaluated for homogeneity by ELISA before use in subsequent assays. Briefly, microtiter plates were coated with 28G4, 16A4, or 2B11 MAb (5.0 μg/ml), and wells were blocked with 5% skim milk and then incubated with affinity-purified IgG (1.0 μg/ml). Bound IgGs were detected with HRP-conjugated goat anti-llama IgG (0.1 μg/ml).
PRNT assay.
WNV-NY1999 stocks were grown in Vero cells and frozen at −80°C until use. Plaque-reduction neutralization titer (PRNT) assays were performed using Vero cell monolayers grown in six-well culture plates at 37°C in 5% CO2. Twofold serial dilutions of heat-inactivated (60°C, 5 min) alpaca serum or affinity-purified IgGs were mixed with an equal volume of WNV (1 × 103 to 2 × 103 PFU per ml), with or without 5% guinea pig complement (Colorado Serum Company). Each monolayer was incubated for 1 h with 100 μl of the virus-antibody mixture, and then monolayers were overlaid with minimum essential medium (MEM; Gibco), 10% fetal bovine serum, 2 mM l-glutamine, 2× penicillin-streptomycin, and 1× ciprofloxacin containing 2% (wt/vol) low-melting-point agarose. Plates were stained on day 3 with neutral red, and plaques were enumerated on day 4. Plaque counts were expressed as percentages of the number of plaques obtained in the absence of serum or antibodies. A reduction of 90% (PRNT90%) or greater was set as the positive threshold.
Preparation of monocyte-derived cell cultures.
A protocol described by Davis et al. (7) was modified to recover mononuclear cells from alpaca blood. Heparinized blood was diluted 1:3 in sterile PBS and layered onto density gradients (upper gradient, Histopaque 1077; lower gradient, Histopaque 1119) (Sigma) in 50-ml polypropylene tubes. Tubes were centrifuged at 700 × g in a swinging-bucket rotor with the brake off for 1 h at ambient temperature. Mononuclear cells were collected from the interface between the plasma and the upper gradient. Leukocyte populations were washed three times by being suspended in PBS and centrifuged (250 × g) for 10 min at 10°C. Viability was estimated with trypan blue, using a hemocytometer, and cellular composition was evaluated by flow cytometry (FACSCalibur; BD Biosciences).
Mononuclear cells were cultured according to the protocol described by Saldarriaga et al. (24), with a few modifications. Pelleted cells were resuspended to a final concentration of 2 × 106 cells/ml (containing approximately 4 × 105 monocytes/ml) in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% heat-inactivated fetal calf serum (Atlanta Biologicals), 0.5 mg/ml gentamicin sulfate (Cellgro), 0.25 μg/ml amphotericin B (Fungizone), 100 U/ml penicillin, 0.1 mg/ml streptomycin, 2 mM l-glutamine, and 50 μM β-mercaptoethanol (Sigma). Cell suspensions (1 ml/well) were added to 24-well culture plates and incubated at 37°C with 5% CO2 for 24 h. Nonadherent cells were removed, and cultures were maintained by replacing the medium every 4 days. Experiments were performed with cultures containing 1 × 105 adherent macrophages per well.
To confirm that adherent cells were macrophage-like, 8-day-old cultures were incubated for 5 min with cold trypsin-EDTA (Sigma) to release cells from the surface. Cytologic preparations (Cytospin) were stained with an α-naphthyl acetate esterase kit (Sigma) following the manufacturer's instructions. Coverslips were mounted with Glycergel (DakoCytomation), and slides were examined using a BX51 microscope fitted with a DP-12 digital camera system (Olympus).
Fc receptor-binding assay.
Purified IgGs (1.0 mg/ml) were dialyzed against PBS (pH 8.5) and incubated with Alexa Fluor 488 (Molecular Probes) (0.1 mg/ml) overnight at 4°C with continuous stirring. Excess dye was removed by washing samples three times with PBS (pH 7.0), using Centricon centrifugal filter devices (YM-30; Millipore) spun at 5,000 × g. The degree of labeling was calculated to be between 4 and 9 mol of dye per mol of IgG. Conjugated IgG preparations were stored at 4°C with 0.1% sodium azide protected from light.
Peripheral blood leukocytes were obtained from alpaca blood by using the protocol described above, except that a single density gradient medium (Histopaque 1119) was used. The experiment was conducted on ice, using cold buffers and reagents. Cells were washed with PBS and then blocked for 1 h with PBS containing 0.1% bovine serum albumin (BSA) and 5% normal goat serum. Cells were subsequently incubated for 1 h with 50 μg/ml Alexa Fluor 488-conjugated IgG1, IgG2, or IgG3 alone or in the presence of HRP-conjugated goat anti-llama IgG at 100, 50, 25, 12.5, or 6.25 μg/ml. Cells were then washed, resuspended in PBS containing 1.0% BSA, and evaluated using flow cytometry. Data were analyzed using the FlowJo (TreeStar) software application.
Antibody-dependent enhancement assay.
Virus (1 × 104 to 2 × 104 PFU) was incubated for 1 h with affinity-purified IgG. Macrophages were incubated with virus-antibody mixtures (multiplicity of infection [MOI] = 0.1) or with medium or virus only as a control. Samples were tested in triplicate. After 3 h, media were aspirated from individual wells, and cells were washed three times with warm (37°C) PBS to remove unbound virus and Abs. Fresh medium was added to each well, and plates were incubated for an additional 36 h at 37°C. Supernatant and cells were harvested separately from each well for estimation of released versus cell-associated virus. Cells were lysed by three cycles of freeze-thawing to release cell-associated virus. Plaque assays were performed as described above by inoculating Vero cells with 200 μl of serially diluted supernatant or lysate.
RESULTS
IgG isotypes induced during natural virus infection.
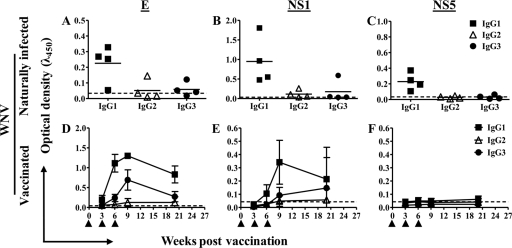
We first assessed the IgG isotypes that were induced during natural infection with WNV. Sera from alpacas (n = 4) that demonstrated serum neutralization (SN) titers ranging from 181 to 1,448 were tested for binding to WNV E, NS1, and NS5 proteins. For three of four animals, the IgG profiles were similar for all antigens tested: specific IgG1 was present, while IgG2 and IgG3 did not exceed the calculated limit of detection of the assay (Fig. 1A to C). In the fourth animal's serum, E- and NS1-specific IgG2 and IgG3 were detected. Such variation may be expected because these samples were acquired during a serological surveillance program for WNV, and it is likely that the histories of the animals varied greatly with regard to exposure to the virus.
FIG. 1.
Antibody isotypes induced by viral antigens. (A to C) WNV-specific IgG isotypes detected in sera of naturally infected alpacas by ELISA, using envelope (E) (A), nonstructural protein NS1 (B), and nonstructural protein NS5 (C). Horizontal bars represent mean OD values per group (n = 4). (D to F) WNV-specific IgG isotypes detected in sera of alpacas vaccinated three times (arrowheads) with killed, adjuvanted WNV. Antibodies specific for E protein (D), NS1 (E), or NS5 (F) were detected by ELISA in sera diluted 1:100. Bars represent standard deviations (SD) of the means (n = 8). Dashed, horizontal lines represent the limit of detection of the assay, calculated as the mean plus three times the average of the SD of the conjugate control.
IgGs induced by WNV vaccine.
We tested serum samples obtained from alpacas that had been administered three doses of a formalin-inactivated WNV vaccine under controlled, experimental conditions (16). All of the animals generated measurable SN titers (45 to 4,096) and produced IgG antibodies specific for WNV E and NS1 proteins, but not NS5, after two doses of vaccine (Fig. 1D to F). Envelope protein-specific IgG3 and IgG1 were induced by vaccination. The response to NS1 proteins was similar but weaker (Fig. 1E). A very poor IgG2 response to NS1 was detected, and overall, the data show that IgG2 was not efficiently induced by vaccination with killed virus. As expected, there were no antibodies induced against the NS5 protein, an RNA polymerase that is absent from the vaccine (Fig. 1F).
Anamnestic response in vaccinated alpacas.
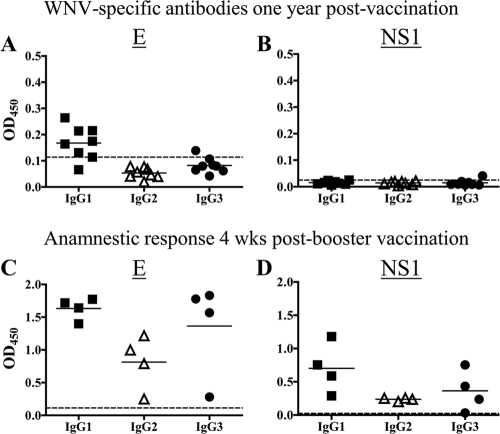
IgGs specific for WNV E protein were detectable 1 year after vaccination (Fig. 2A); however, anti-NS1 IgGs were not (Fig. 2B). Administration of vaccine at this time elicited an anamnestic response that included IgG1 and IgG3 specific for the E and NS1 proteins (Fig. 2C and D). In addition, a marked increase in IgG2 against the E and NS1 proteins was detected. These results document that alpacas were primed by vaccination to produce all three IgG isotypes.
FIG. 2.
Detection of circulating antibodies and induction of anamnestic response against WNV E and NS1 proteins at 1 year postvaccination. (A and B) Anti-WNV antibodies in alpacas (n = 8) at 1 year postvaccination. (C and D) Anamnestic response at 4 weeks post-booster vaccination (n = 4 alpacas). Antibodies were detected by ELISA in sera diluted 1:100. Horizontal bars represent mean OD values per group. Dashed, horizontal lines represent the limit of detection of the assay, calculated as described in the legend to Fig. 1.
Evaluation of virus-neutralizing activities of alpaca IgGs.
The host cell receptor-binding domains of WNV are located within the E protein (5), and antibodies elicited against this protein neutralize the virus. Since both HCAbs and conventional IgG were induced against the E protein, we next sought to compare their efficiencies in neutralization of WNV.
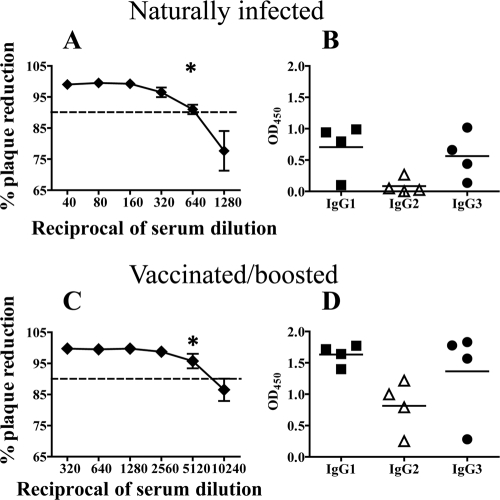
To quantify neutralizing activities of WNV-specific antibodies, we compared the PRNTs of sera from naturally infected versus vaccinated/boosted alpacas. Sera from vaccinated/boosted alpacas were eightfold more potent in PRNT90% assays than sera from naturally infected animals (Fig. 3A and C). Since sera likely contained highly efficient, neutralizing IgM antibodies and we wanted to determine whether IgGs participated in neutralization, we tested IgG1, IgG2, and IgG3 purified from sera by using affinity columns prepared with isotype-specific MAbs. Antibodies were purified from sera of four naturally infected and four vaccinated/boosted alpacas. The homogeneity of each IgG preparation was confirmed by sandwich ELISA (data not shown). Before testing the purified antibodies in PRNT assays, we confirmed that they had not deteriorated during the purification process by testing their binding to WNV E proteins in ELISA. Binding activities were preserved (Fig. 3B and D), with the exception that binding of IgG3 from naturally infected alpacas improved substantially after purification (Fig. 3B versus Fig. 1A), perhaps because the larger conventional antibodies hindered access of IgG3 to binding sites on the E protein.
FIG. 3.
Binding activities of antibodies in sera from naturally infected and vaccinated/boosted alpacas. Sera were collected at the onset of clinical signs for naturally exposed alpacas and at 1 month postboosting for vaccinated/boosted alpacas. (A and C) Detection of neutralizing antibodies in sera from naturally infected (A) and vaccinated/boosted (C) alpacas. A standard PRNT assay was conducted with sera in the presence of complement (n = 4 alpacas per group). The asterisk indicates the end point. Dashed, horizontal lines represent the PRNT90%, which was set as the positive threshold. (B and D) WNV E protein-specific IgGs detected in sera from naturally infected (B) and vaccinated/boosted (D) alpacas. IgGs were affinity purified from sera and then assayed for binding to WNV E protein in ELISA. Horizontal bars represent mean OD values per group.
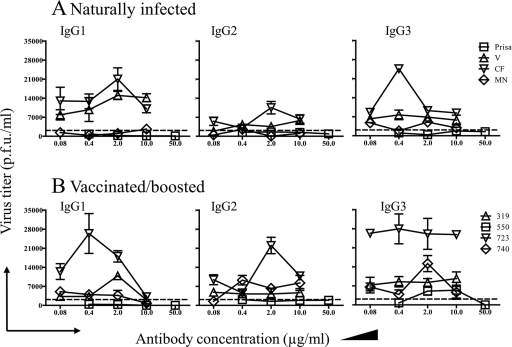
IgG1 and IgG3 induced by either natural exposure or vaccination demonstrated potent neutralizing activities (Fig. 4). Conversely, IgG2 induced by vaccination demonstrated suboptimal neutralization (plaque reduction of <90%) (Fig. 4B). Neutralization by IgG1, IgG2, and IgG3 was not dependent upon complement, although the addition of complement to the assay enhanced neutralization by all three isotypes (not shown).
FIG. 4.
Neutralization of WNV by IgGs purified from alpaca sera. PRNT assays were conducted on Vero cells, using affinity-purified IgGs obtained from naturally infected (A) and vaccinated/boosted (B) alpacas. Dashed, horizontal lines represent the PRNT90%, which was set as the positive threshold. Symbols are labeled with names or numbers assigned to the alpacas from which serum samples were obtained.
Demonstration of IgG binding to mononuclear leukocytes.
Binding to FcR on leukocytes is an important effector function of IgG. We employed flow cytometry to determine whether conventional or HC isotypes bind receptors displayed on the surfaces of mononuclear cells.
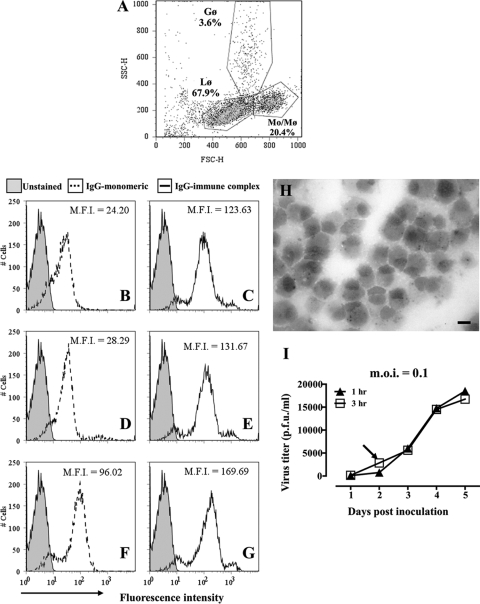
Monocytes were gated based on forward versus side scatter (Fig. 5A). Increased fluorescence intensities of cells incubated with fluorochrome-conjugated IgGs illustrated that all three isotypes were able to bind to surfaces of monocytes (Fig. 5B, D, and F). No effort was made to remove aggregates from the labeled antibody preparations, so it was not determined whether binding was via monomeric or aggregated IgG. Aggregation with goat anti-llama enhanced binding of all three isotypes to monocytes (Fig. 5C, E, and G). Unlabeled goat IgG was included in the blocking solution to reduce complex binding to monocytes via the goat antibodies. Taken together, the results provide evidence that alpaca monocytes express receptors for conventional and HC IgGs.
FIG. 5.
Antibody binding and WNV infection of cultured alpaca peripheral blood monocytes. (A) Flow cytometric scatter plot (forward versus side scatter) of the low-density cell fraction recovered from density gradients. Gates were set around lymphocytes (Lφ), monocytes (Mo/Mφ), and granulocytes (Gφ). The representation of each cell type within the population is indicated. (B to G) Binding of purified fluorescent alpaca IgGs to monocytes and macrophages. Leukocytes were incubated on ice with Alexa Fluor 488-conjugated IgG1 (B), IgG2 (D), or IgG3 (F) alone or with HRP-conjugated goat anti-llama IgG to form immune complexes (C, E, and G). Untreated cells were used as controls. Binding of fluorochrome-labeled IgGs to cells was assessed by flow cytometry. Histograms of fluorescence intensities demonstrate the binding of IgGs to cells within the Mo/Mφ gate shown in panel A. M.F.I., mean fluorescence intensity. (H) Cytospin preparation of adherent cells from low-density fraction after 8 days in culture. Macrophages containing cytoplasmic esterase specific for α-naphthyl acetate stain black. Bar, 10 μm. (I) Macrophages (1 × 105) were incubated with WNV (MOI = 0.1) for 1 h (filled symbols) or 3 h (open symbols), washed with PBS, and cultured at 37°C for 5 days. Supernatant was sampled daily, and virus numbers were estimated by plaque assay. The arrow indicates the inoculation period (3 h) and time point of supernatant collection used in subsequent assays.
Effect of IgG on WNV infection of macrophages.
To assess the effect of alpaca IgGs on virus replication in macrophages, we conducted experiments in which alpaca macrophages were inoculated with WNV in the presence of purified IgGs.
Macrophages were prepared from alpaca peripheral blood leukocytes. Monocytes were recovered by density gradient centrifugation (Fig. 5A) and were further enriched in culture by selecting the adherent cell population. Within 8 days of culture, adherent cells had differentiated into macrophage-like cells that expressed cytoplasmic esterases specific for α-naphthyl acetate (Fig. 5H).
Inoculation of cultures with virus at an MOI of 0.1 resulted in replication of WNV. Virus titers were low in supernatants (20,000 PFU/ml) and cell lysates (300 PFU/ml) collected within 5 days of inoculation, suggesting that alpaca macrophages are not highly permissive to WNV infection (Fig. 5I) compared to bone marrow-derived macrophages from mice (25). The results observed were not affected by inoculation time (Fig. 5I) or the addition of complement (not shown).
To assess the effects of IgGs on virus replication in macrophages, WNV was incubated with IgG at concentrations at or below the 90% neutralization threshold in Vero cells (10, 2, 0.4, and 0.08 μg/ml [n = 3 alpacas] or 50 μg/ml [n = 1 alpaca {Prisa}]), with or without added complement, prior to inoculation of macrophages with virus (Fig. 6 and data not shown). Cells (1 × 105) were incubated for 3 h with 1 × 104 PFU of WNV (MOI = 0.1), and supernatants were collected 36 h later. Enhancement of infectivity was observed with all three isotypes but was variable among the preparations tested, including those giving comparable results on Vero cells. In some instances, virus replication was enhanced >10-fold by IgG1 and IgG3 (animals V and 723). In other cases, little or no enhancement was observed (Prisa and animal 550) (Fig. 6A and B), demonstrating that enhancement was not mediated by antibodies in a nonspecific manner. Altogether, antibodies from seven of the eight animals showed enhancement with at least one isotype. Inclusion of complement reduced the enhancement effects in 21 of the 24 preparations evaluated (i.e., 6/8 preparations of IgG1, 8/8 preparations of IgG2, and 7/8 preparations of IgG3) (data not shown). Titers of cell-associated viruses correlated with those of released viruses but were considerably reduced (data not shown).
FIG. 6.
Antibody-dependent enhancement of WNV replication in alpaca macrophages. Virus was incubated with IgGs purified from naturally infected (A) or vaccinated/boosted (B) animal sera at concentrations that yielded less than 90% neutralization in PRNT assays. Virus-antibody mixtures were inoculated onto monolayers of alpaca macrophages, and supernatants were collected at 48 h postinoculation. Virus released from infected cells was quantified by plaque assay. Dashed lines represent virus obtained from macrophages inoculated with WNV in the absence of antibody. Symbols are labeled with names or numbers assigned to the alpacas from which serum samples were obtained.
Overall, the data are compatible with the conclusion that WNV infection of camelid macrophages is inefficient; however, infectivity can be enhanced by low concentrations of WNV-specific IgG antibodies. In the presence of complement, any enhancement was abolished.
DISCUSSION
Clearance of pathogens by antibodies is initiated when an antigenic site is bound by the CDRs of the immunoglobulin V domain. Binding may neutralize viruses, toxins, or enzymes deployed by the pathogen. While published reports document the extraordinary ability of camelid HCAbs to inhibit enzymes by inserting an extended CDR into the active site (4, 17, 28), a very limited volume of published work describes the specificity and function of HCAbs in the context of infection. In one study, HCAbs produced by camels in response to infection with trypanosomes displayed a broad repertoire of antigen specificities (2, 13). In another report, llama HCAbs were shown to display a more restricted and, in some ways, distinctive repertoire of specificities for bacterial antigens than those of conventional IgGs (30). To our knowledge, there are no reports describing the specificities or functions of camelid IgGs produced in response to vaccination or infection with viruses. Such studies have been hindered by the lack of well-characterized, isotype-specific reagents. The availability of MAbs specific for llama and alpaca IgG1, IgG2, and IgG3 (6) has enabled us to investigate specificities and regulation and effector functions of HCAbs in antiviral immunity.
WNV is related to flaviviruses that cause dengue fever, yellow fever, and Japanese encephalitis (18). Its 11-kb RNA genome encodes a single polyprotein that is cleaved by both viral and host enzymes to yield three structural and seven nonstructural proteins (3). The E protein of WNV incorporates putative receptor-binding sites, and antibodies raised against E protein neutralize the virus (1, 23, 27, 31, 32, 35). A successful humoral immune response against virus infection is determined by the ability of the elicited antibodies to bind and neutralize virus. In our studies, natural infection with WNV induced E-specific IgG3 and conventional IgG1. IgG3 exhibited neutralizing activity that was comparable to that of IgG1. Vaccination with inactivated virus also induced neutralizing IgG1 and IgG3. In contrast, IgG2 specific for E protein was detected only in the anamnestic response to vaccination and, when present, was not potent in neutralization assays. The basis for this failure is not clear, and we are cautious in interpreting the result because the number of animals studied was limited. Alpaca IgG2 and IgG3 differ in mass (6) and may also differ in structure. Camel HCAbs are known to vary in hinge length (14). Such variation may alter molecular flexibility and thereby influence neutralizing activity.
Antigen binding enables antibodies to perform effector functions. Activation of the complement pathway, opsonization of pathogens, activation of mast cells, and the induction of antibody-dependent cellular cytotoxicity (ADCC) are critical in host defense against pathogens. Cell-dependent effector functions of IgG are mediated by Fc receptors (FcγRs) on leukocytes. FcγRs vary in affinity and specificity for different isotypes (22). Effector functions and leukocyte binding by camelid IgGs have not been described. We found that both conventional and HC isotypes bound to the surfaces of blood monocytes. We did not determine whether the binding was via high- or low-affinity receptors, a question that merits further investigation. In addition, virus neutralization by HCAbs was enhanced in the presence of complement, indicating that they also possess complement binding as an effector function.
Antibody-dependent enhancement (ADE) of viral infection is a phenomenon that is characteristic of flaviviruses. This process is mediated by IgG antibodies, and although there are several mechanisms proposed to explain it, the most widely accepted one involves Fc receptors (13). Since it has been established that IgGs mediate ADE of West Nile virus infection (15), we asked whether camelid IgGs could mediate ADE. In contrast to equine cells (12), alpaca macrophages incubated with virus, alone or together with complement, were not highly permissive to WNV replication. When immune complexes were formed by incubating the virus with subneutralizing concentrations of purified IgG, ADE was demonstrable for all three isotypes. Consistent with these observations, flow cytometric analysis revealed that each isotype bound to the surfaces of macrophages. The data support the conclusion that HCAbs and conventional IgG bind putative FcγRs on alpaca macrophages. To our knowledge, FcγRs have not yet been described for camelid leukocytes; however, IgG2 CH domains incorporate amino acid motifs that are known to be essential for binding of human IgG to FcγRI (20).
In aggregate, our data provide evidence that the alpaca HC isotypes, i.e., IgG2 and IgG3, differ in both regulation and function. Virus-specific IgG2 was not efficiently induced by either vaccination or infection with WNV. When induced during an anamnestic response to vaccination, IgG2 did not efficiently neutralize WNV. In contrast, IgG3 was induced by both infection and vaccination and was potent in neutralization assays. Conventional IgG1 dominated all of the immune responses examined. All three isotypes were capable of binding to and promoting infection of macrophages by WNV via an ADE-like mechanism. The results enrich our understanding of camelid immune defense and provide a foundation of knowledge that will be useful in the development of effective vaccines and diagnostic tests for these animals.
Acknowledgments
We thank Diana Meskill, Lisa Blum, Lucille Gagliardo, Joseph Lembo, and Kate Justus for assistance with cell processing.
This work was supported by a grant (to J.A.A.) from the Program for Collaborative Research in Preclinical and Clinical Sciences and by a Dean's Diversity Award (to L.P.D.), College of Veterinary Medicine, Cornell University. M.A.K.'s research was supported by the Alpaca Research Foundation.
Footnotes
Published ahead of print on 2 December 2009.
REFERENCES
- 1.Beasley, D. W., and A. D. Barrett. 2002. Identification of neutralizing epitopes within structural domain III of the West Nile virus envelope protein. J. Virol. 76:13097-13100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bjorck, L., and G. Kronvall. 1984. Purification and some properties of streptococcal protein G, a novel IgG-binding reagent. J. Immunol. 133:969-974. [PubMed] [Google Scholar]
- 3.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 4.Chan, P. H., E. Pardon, L. Menzer, E. De Genst, J. R. Kumita, J. Christodoulou, D. Saerens, A. Brans, F. Bouillenne, D. B. Archer, C. V. Robinson, S. Muyldermans, A. Matagne, C. Redfield, L. Wyns, C. M. Dobson, and M. Dumoulin. 2008. Engineering a camelid antibody fragment that binds to the active site of human lysozyme and inhibits its conversion into amyloid fibrils. Biochemistry 47:11041-11054. [DOI] [PubMed] [Google Scholar]
- 5.Chu, J. J., R. Rajamanonmani, J. Li, R. Bhuvanakantham, J. Lescar, and M. L. Ng. 2005. Inhibition of West Nile virus entry by using a recombinant domain III from the envelope glycoprotein. J. Gen. Virol. 86:405-412. [DOI] [PubMed] [Google Scholar]
- 6.Daley, L. P., L. F. Gagliardo, M. S. Duffy, M. C. Smith, and J. A. Appleton. 2005. Application of monoclonal antibodies in functional and comparative investigations of heavy-chain immunoglobulins in new world camelids. Clin. Diagn. Lab. Immunol. 12:380-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis, W. C., L. R. Heirman, M. J. Hamilton, S. M. Parish, G. M. Barrington, A. Loftis, and M. Rogers. 2000. Flow cytometric analysis of an immunodeficiency disorder affecting juvenile llamas. Vet. Immunol. Immunopathol. 74:103-120. [DOI] [PubMed] [Google Scholar]
- 8.Decanniere, K., A. Desmyter, M. Lauwereys, M. A. Ghahroudi, S. Muyldermans, and L. Wyns. 1999. A single-domain antibody fragment in complex with RNase A: non-canonical loop structures and nanomolar affinity using two CDR loops. Structure 7:361-370. [DOI] [PubMed] [Google Scholar]
- 9.Decanniere, K., S. Muyldermans, and L. Wyns. 2000. Canonical antigen-binding loop structures in immunoglobulins: more structures, more canonical classes? J. Mol. Biol. 300:83-91. [DOI] [PubMed] [Google Scholar]
- 10.Desmyter, A., K. Decanniere, S. Muyldermans, and L. Wyns. 2001. Antigen specificity and high affinity binding provided by one single loop of a camel single-domain antibody. J. Biol. Chem. 276:26285-26290. [DOI] [PubMed] [Google Scholar]
- 11.Desmyter, A., T. R. Transue, M. A. Ghahroudi, M. H. Thi, F. Poortmans, R. Hamers, S. Muyldermans, and L. Wyns. 1996. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat. Struct. Biol. 3:803-811. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Tapia, D., C. M. Loiacono, and S. B. Kleiboeker. 2006. Replication of West Nile virus in equine peripheral blood mononuclear cells. Vet. Immunol. Immunopathol. 110:229-244. [DOI] [PubMed] [Google Scholar]
- 13.Halstead, S. B., E. J. O'Rourke, and A. C. Allison. 1977. Dengue viruses and mononuclear phagocytes. II. Identity of blood and tissue leukocytes supporting in vitro infection. J. Exp. Med. 146:218-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamers-Casterman, C., T. Atarhouch, S. Muyldermans, G. Robinson, C. Hamers, E. B. Songa, N. Bendahman, and R. Hamers. 1993. Naturally occurring antibodies devoid of light chains. Nature 363:446-448. [DOI] [PubMed] [Google Scholar]
- 15.Hawkes, R. A. 1964. Enhancement of the infectivity of arboviruses by specific antisera produced in domestic fowls. Aust. J. Exp. Biol. Med. Sci. 42:465-482. [DOI] [PubMed] [Google Scholar]
- 16.Kutzler, M. A., R. J. Baker, and D. E. Mattson. 2004. Humoral response to West Nile virus vaccination in alpacas and llamas. J. Am. Vet. Med. Assoc. 225:414-416. [DOI] [PubMed] [Google Scholar]
- 17.Lauwereys, M., M. Arbabi Ghahroudi, A. Desmyter, J. Kinne, W. Holzer, E. De Genst, L. Wyns, and S. Muyldermans. 1998. Potent enzyme inhibitors derived from dromedary heavy-chain antibodies. EMBO J. 17:3512-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukhopadhyay, S., R. J. Kuhn, and M. G. Rossmann. 2005. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3:13-22. [DOI] [PubMed] [Google Scholar]
- 19.Muyldermans, S., C. Cambillau, and L. Wyns. 2001. Recognition of antigens by single-domain antibody fragments: the superfluous luxury of paired domains. Trends Biochem. Sci. 26:230-235. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen, V. K., R. Hamers, L. Wyns, and S. Muyldermans. 1999. Loss of splice consensus signal is responsible for the removal of the entire C(H)1 domain of the functional camel IgG2A heavy-chain antibodies. Mol. Immunol. 36:515-524. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen, V. K., S. Muyldermans, and R. Hamers. 1998. The specific variable domain of camel heavy-chain antibodies is encoded in the germline. J. Mol. Biol. 275:413-418. [DOI] [PubMed] [Google Scholar]
- 22.Nimmerjahn, F., and J. V. Ravetch. 2005. Divergent immunoglobulin G subclass activity through selective Fc receptor binding. Science 310:1510-1512. [DOI] [PubMed] [Google Scholar]
- 23.Oliphant, T., M. Engle, G. E. Nybakken, C. Doane, S. Johnson, L. Huang, S. Gorlatov, E. Mehlhop, A. Marri, K. M. Chung, G. D. Ebel, L. D. Kramer, D. H. Fremont, and M. S. Diamond. 2005. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat. Med. 11:522-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saldarriaga, O. A., J. I. Velaquez, J. E. Ossa, and M. T. Rugeles. 2003. Standardization of bovine macrophage monolayers and isolation and culture of trypanosomes. Mem. Inst. Oswaldo Cruz 98:269-271. [DOI] [PubMed] [Google Scholar]
- 25.Shrestha, B., B. Zhang, W. E. Purtha, R. S. Klein, and M. S. Diamond. 2008. Tumor necrosis factor alpha protects against lethal West Nile virus infection by promoting trafficking of mononuclear leukocytes into the central nervous system. J. Virol. 82:8956-8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spinelli, S., L. Frenken, D. Bourgeois, L. de Ron, W. Bos, T. Verrips, C. Anguille, C. Cambillau, and M. Tegoni. 1996. The crystal structure of a llama heavy chain variable domain. Nat. Struct. Biol. 3:752-757. [DOI] [PubMed] [Google Scholar]
- 27.Thompson, B. S., B. Moesker, J. M. Smit, J. Wilschut, M. S. Diamond, and D. H. Fremont. 2009. A therapeutic antibody against West Nile virus neutralizes infection by blocking fusion within endosomes. PLoS Pathog. 5:e1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Transue, T. R., E. De Genst, M. A. Ghahroudi, L. Wyns, and S. Muyldermans. 1998. Camel single-domain antibody inhibits enzyme by mimicking carbohydrate substrate. Proteins 32:515-522. [DOI] [PubMed] [Google Scholar]
- 29.Ungar-Waron, H., E. Elias, A. Gluckman, and Z. Trainin. 1987. Dromedary IgG: purification, characterization, and quantification in sera of dams and newborns. Isr. J. Vet. Med. 43:198-203. [Google Scholar]
- 30.van der Linden, R., B. de Geus, W. Stok, W. Bos, D. van Wassenaar, T. Verrips, and L. Frenken. 2000. Induction of immune responses and molecular cloning of the heavy chain antibody repertoire of Lama glama. J. Immunol. Methods 240:185-195. [DOI] [PubMed] [Google Scholar]
- 31.Vogt, M. R., B. Moesker, J. Goudsmit, M. Jongeneelen, S. K. Austin, T. Oliphant, S. Nelson, T. C. Pierson, J. Wilschut, M. Throsby, and M. S. Diamond. 2009. Human monoclonal antibodies against West Nile virus induced by natural infection neutralize at a postattachment step. J. Virol. 83:6494-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volk, D. E., D. W. Beasley, D. A. Kallick, M. R. Holbrook, A. D. Barrett, and D. G. Gorenstein. 2004. Solution structure and antibody binding studies of the envelope protein domain III from the New York strain of West Nile virus. J. Biol. Chem. 279:38755-38761. [DOI] [PubMed] [Google Scholar]
- 33.Vu, K. B., M. A. Ghahroudi, L. Wyns, and S. Muyldermans. 1997. Comparison of llama VH sequences from conventional and heavy chain antibodies. Mol. Immunol. 34:1121-1131. [DOI] [PubMed] [Google Scholar]
- 34.Woolven, B. P., L. G. Frenken, P. van der Logt, and P. J. Nicholls. 1999. The structure of the llama heavy chain constant genes reveals a mechanism for heavy-chain antibody formation. Immunogenetics 50:98-101. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, S., M. R. Vogt, T. Oliphant, M. Engle, E. I. Bovshik, M. S. Diamond, and D. W. Beasley. 2009. Development of resistance to passive therapy with a potently neutralizing humanized monoclonal antibody against West Nile virus. J. Infect. Dis. 200:202-205. [DOI] [PMC free article] [PubMed] [Google Scholar]