Abstract
Although 16S rRNA gene sequence analysis is employed most often for the definitive identification of Nocardia species, alternate molecular methods and polymorphisms in other gene targets have also enabled species determinations. We evaluated a combined Nocardia PCR-based reverse line blot (RLB) hybridization assay based on 16S and 16S-23S rRNA gene spacer region polymorphisms to identify 12 American Type Culture Collection and 123 clinical Nocardia isolates representing 14 species; results were compared with results from 16S rRNA gene sequencing. Thirteen 16S rRNA gene-based (two group-specific and 11 species-specific) and five 16S-23S spacer-targeted (two taxon-specific and three species-specific) probes were utilized. 16S rRNA gene-based probes correctly identified 124 of 135 isolates (sensitivity, 92%) but were unable to identify Nocardia paucivorans strains (n = 10 strains) and a Nocardia asteroides isolate with a novel 16S rRNA gene sequence. Nocardia farcinica and Nocardia cyriacigeorgica strains were identified by the sequential use of an N. farcinica-“negative” probe and a combined N. farcinica/N. cyriacigeorgica probe. The assay specificity was high (99%) except for weak cross-reactivity between the Nocardia brasiliensis probe with the Nocardia thailandica DNA product; however, cross-hybridization with closely related nontarget species may occur. The incorporation of 16S-23S rRNA gene spacer-based probes enabled the identification of all N. paucivorans strains. The overall sensitivity using both probe sets was >99%. Both N. farcinica-specific 16S-23S rRNA gene spacer-directed probes were required to identify all N. farcinica stains by using this probe set. The study demonstrates the utility of a combined PCR/RLB assay for the identification of clinically relevant Nocardia species and its potential for studying subtypes of N. farcinica. Where species assignment is ambiguous or not possible, 16S rRNA gene sequencing is recommended.
Nocardia species are ubiquitous environmental bacteria that cause suppurative infections ranging from localized lung involvement to disseminated disease. The identification of clinical isolates to the species level is important to characterize associated disease manifestations, predict antimicrobial susceptibility, and identify differences in epidemiology (5, 21, 26). Because standard phenotype-based identification methods are slow (10 to 21 days), are often imprecise, and cannot identify novel species (5, 21), various nucleic acid amplification tools targeting conserved gene regions have been used to provide rapid, accurate species determinations.
Of these tools, 16S rRNA gene sequence analysis is the most frequently used and has become the “gold standard” method for definitive species identification (5, 7, 9, 25). Sequence polymorphisms within the Nocardia 65-kDa heat shock protein (hsp65) and essential secretary protein A (SecA1) gene (secA1) targets were also reported to enable the identification of isolates to the species level (12, 24, 28). The introduction of these identification methods has predictably led to substantial species reassignment and the reporting of new species within the genus, especially among “Nocardia asteroides” isolates: over 80 species have now been described, ∼33 of which have been implicated in human disease (http://www.ncbi.nlm.nih.gov).
However, the practical limitations of 16S rRNA gene sequencing have been realized (23). This method is often unable to distinguish between certain closely related Nocardia species due to insufficient interspecies polymorphisms within the 16S rRNA gene sequences (5, 9, 19). Misidentifications may also occur as a result of multiple, but different, copies of the 16S rRNA gene in certain species such as Nocardia nova (10, 11). Furthermore, sequencing remains time-consuming. As such, the continuing evaluation of alternate PCR-based methods and/or gene targets to facilitate species identification is important. Formats with diagnostic potential include real-time PCR, restriction enzyme analysis of the 16S rRNA gene or hsp65, and random amplification of polymorphic DNA (1, 5, 17, 24). However, the clinical application of these approaches may be limited by the expanding number of pathogenic species; to date, identification assays have focused largely on the identification of Nocardia farcinica (4, 16), as this species is resistant to many antibiotics used to treat nocardiosis (5, 31). Given that species distribution varies by geographic region (5), the selection of an assay targeting clinically relevant species that are prevalent in a region lends itself as a reasonable approach.
We previously developed a simple method to rapidly detect and identify bacterial and fungal pathogens using PCR followed by hybridization with species-specific oligonucleotides in a reverse line blot (RLB) assay (20, 32, 34). This approach has the capacity for the simultaneous analysis of a large number (∼40) of strains against multiple probes and can be adapted to include different gene targets within the same assay (14). In the present study, we designed and incorporated into an RLB format species- and group-specific probes targeting Nocardia 16S rRNA genes as well as probes directed at the 16S-23S rRNA gene spacer (or intergenic transcribed spacer [ITS]) regions to identify the more common clinically important Nocardia species (5, 8, 24, 27). The selection of target species was based on the known or likely prevalence of the pathogen (5, 19, 22) and where at least one isolate of the species was available for study. We then tested the ability of the RLB assay to identify 135 Nocardia reference and clinical strains in our culture collection and compared the results with 5′-end 606-bp 16S rRNA gene sequencing. 16S-23S rRNA gene spacer-targeted probes were chosen as an adjunct to 16S rRNA gene probes to optimize the sensitivity and specificity of the RLB assay since this locus has shown promise as a target for species identification for other bacteria (29, 33).
MATERIALS AND METHODS
Nocardia organisms.
One hundred thirty-five Nocardia isolates were studied. These comprised 12 American Type Culture Collection (ATCC) (Rockville, MD) strains (Nocardia abscessus ATCC BAA-279T, N. asteroides ATCC 19247T, Nocardia brasiliensis ATCC 19296T, Nocardia brevicatena ATCC 15333T, Nocardia carnea ATCC 6847T, N. farcinica ATCC 3308, N. farcinica ATCC 3318T, Nocardia nova ATCC 33726T, Nocardia otitidiscaviarum ATCC 14629T, Nocardia paucivorans ATCC BAA-278T, Nocardia transvalensis ATCC 6865T, and Nocardia veterana ATCC BAA-509) and 123 clinical isolates obtained from the Centre for Infectious Diseases and Microbiology Laboratory Services, Westmead Hospital, Sydney, Australia (Table 1). Cultured clinical isolates were obtained from a range of body sites from separate patients from 1997 to 2007. All isolates were identified by standard phenotypic methods and antibiotic susceptibility profiles (21). Organisms were cultured in brain heart infusion (BHI) broth (Amyl Media, Dandenong, Australia) for 3 to 15 days at 37°C in air prior to analysis by RLB assay and 16S rRNA gene sequencing (routinely performed in our laboratory since 2005).
TABLE 1.
Species identification of 123 clinical Nocardia isolates by phenotype-based methods, 16S rRNA gene sequencing, and reverse line blot assay
| Strain | Identification bya: |
|||
|---|---|---|---|---|
| Phenotype | 16S rRNA gene sequencing | 16S rRNA gene RLB | 16S-23S rRNA gene spacer-RLB | |
| 00-088-3187 | N. asteroides complex | N. abscessus | N. abscessus | |
| 00-129-3338 | N. asteroides complex | N. paucivorans | NA | N. paucivorans |
| 00-159-1584 | N. asteroides | N. cyriacigeorgica | N. cyriacigeorgica | |
| 00-236-1204 | N. asteroides | N. cyriacigeorgica | N. cyriacigeorgica | |
| 00-279-0751 | N. asteroides complex | N. cyriacigeorgica | N. cyriacigeorgica | |
| 00-355-2649 | N. asteroides | N. thailandica | N. thailandica | |
| 00-361-0960 | N. asteroides | N. cyriacigeorgica | N. cyriacigeorgica | |
| 01-067-1349 | N. asteroides | N. nova | N. nova | |
| 01-226-3924 | N. asteroides | N. cyriacigeorgica | N. cyriacigeorgica | |
| 01-248-3107 | N. asteroides | N. abscessus | N. abscessus | |
| 01-271-2702 | N. asteroides | N. farcinica | N. farcinica | N. farcinica |
| 01-317-3331 | N. asteroides | N. cyriacigeorgica | N. cyriacigeorgica | |
| 02-071-3627 | N. asteroides | N. asteroides | NSb | |
| 04-128-3054 | N. asteroides | N. asteroides | N. asteroides | |
| 02-150-3195 | N. asteroides | N. cyriacigeorgica | N. cyriacigeorgica | |
| 02-337-3608 | N. asteroides | N. cyriacigeorgica | N. cyriacigeorgica | |
| 02-347-2673 | N. asteroides complex | N. abscessus | N. abscessus | |
| 02-361-1282 | N. asteroides | N. cyriacigeorgica | N. cyriacigeorgica | |
| 03-044-3083 | N. asteroides | N. cyriacigeorgica | N. cyriacigeorgica | |
| 03-051-2726 | N. asteroides | N. cyriacigeorgica | N. cyriacigeorgica | |
| 03-065-1348 | N. asteroides | N. cyriacigeorgica | N. cyriacigeorgica | |
| 03-072-1482 | N. asteroides | N. cyriacigeorgica | N. cyriacigeorgica | |
| 03-078-3387 | N. asteroides | N. cyriacigeorgica | N. cyriacigeorgica | |
| 03-090-3765 | N. asteroides | N. cyriacigeorgica | N. cyriacigeorgica | |
| 03-100-2450 | N. asteroides complex | N. paucivorans | NA | N. paucivorans |
| 03-105-2890 | N. asteroides | N. cyriacigeorgica | N. cyriacigeorgica | |
| 03-125-3479 | N. asteroides | N. cyriacigeorgica | N. cyriacigeorgica | |
| 03-141-3073 | N. asteroides complex | N. paucivorans | NA | N. paucivorans |
| 03-185-3304 | N. asteroides | N. paucivorans | NA | N. paucivorans |
| 03-240-2758 | N. asteroides complex | N. paucivorans | NA | N. paucivorans |
| 03-246-1321 | N. asteroides complex | N. cyriacigeorgica | N. cyriacigeorgica | |
| 03-336-3513 | N. asteroides | N. cyriacigeorgica | N. cyriacigeorgica | |
| 03-342-3119 | N. asteroides | N. cyriacigeorgica | N. cyriacigeorgica | |
| 04-124-4049 | N. asteroides complex | N. nova | N. nova | |
| 04-141-1037 | N. asteroides complex | N. farcinica | N. farcinica | N. farcinica |
| 04-167-1813 | N. asteroides | N. cyriacigeorgica | N. cyriacigeorgica | |
| 04-181-3939 | N. asteroides | N. cyriacigeorgica | N. cyriacigeorgica | |
| 04-204-3567 | N. asteroides | N. veterana | N. veterana | |
| 05-034-0715 | N. asteroides complex | N. cyriacigeorgica | N. cyriacigeorgica | |
| 05-040-4253 | N. asteroides complex | N. cyriacigeorgica | N. cyriacigeorgica | |
| 05-054-3687 | N. asteroides | N. veterana | N. veterana | |
| 05-062-1428 | N. asteroides complex | N. cyriacigeorgica | N. cyriacigeorgica | |
| 05-073-0700 | N. asteroides complex | N. cyriacigeorgica | N. cyriacigeorgica | |
| 05-111-2308 | N. asteroides complex | N. cyriacigeorgica | N. cyriacigeorgica | |
| 05-133-3591 | N. asteroides complex | N. cyriacigeorgica | N. cyriacigeorgica | |
| 05-144-3166 | N. asteroides complex | N. veterana | N. veterana | |
| 05-318-3826 | N. asteroides complex | N. cyriacigeorgica | N. cyriacigeorgica | |
| 05-342-2208 | N. asteroides complex | N. cyriacigeorgica | N. cyriacigeorgica | |
| 07-154-2262 | Not done | N. cyriacigeorgica | N. cyriacigeorgica | |
| 07-296-2914 | Not done | N. cyriacigeorgica | N. cyriacigeorgica | |
| 01-114-2816 | N. brasiliensis | N. otitidiscaviarum | N. otitidiscaviarum | |
| 01-255-3190 | N. brasiliensis | N. brasiliensis | N. brasiliensis | |
| 02-151-1206 | N. brasiliensis | N. brasiliensis | N. brasiliensis | |
| 03-174-4234 | N. brasiliensis | N. brasiliensis | N. brasiliensis | |
| 03-273-2825 | N. brasiliensis | N. brasiliensis | N. brasiliensis | |
| 04-105-2324 | N. brasiliensis | N. brasiliensis | N. brasiliensis | |
| 05-035-2843 | N. brasiliensis | N. brasiliensis | N. brasiliensis | |
| 06-158-4578 | N. brasiliensis | N. brasiliensis | N. brasiliensis | |
| 07-039-0480 | N. brasiliensis | N. brasiliensis | N. brasiliensis | |
| 00-122-3261 | N. farcinica | N. farcinica | N. farcinica | N. farcinica |
| 01-109-2248 | N. farcinica | N. farcinica | N. farcinica | N. farcinica |
| 02-266-1081 | N. farcinica | N. farcinica | N. farcinica | N. farcinica |
| 02-330-2412 | N. farcinica | N. farcinica | N. farcinica | N. farcinica |
| 03-008-1568 | N. farcinica | N. farcinica | N. farcinica | N. farcinica |
| 03-118-3883 | N. farcinica | N. farcinica | N. farcinica | N. farcinica |
| 03-225-1079 | N. farcinica | N. cyriacigeorgica | N. cyriacigeorgica | |
| 05-118-2483 | N. farcinica | N. farcinica | N. farcinica | N. farcinica |
| 05-048-3600 | N. farcinica | N. farcinica | N. farcinica | N. farcinica |
| 05-186-4091 | N. farcinia | N. farcinica | N. farcinica | N. farcinica |
| 05-207-0815 | N. farcinica | N. farcinica | N. farcinica | N. farcinica |
| 05-249-2502 | N. farcinica | N. farcinica | N. farcinica | N. farcinica |
| 06-280-1688 | N. farcinica | N. farcinica | N. farcinica | N. farcinica |
| 07-023-0418 | N. farcinica | N. farcinica | N. farcinica | N. farcinica |
| 07-226-2287 | N. farcinica | N. farcinica | N. farcinica | N. farcinica |
| 07-247-4763 | N. farcinica | N. farcinica | N. farcinica | N. farcinica |
| 98-286-1121 | N. nova | N. nova | N. nova | |
| 99-270-2693 | N. nova | N. nova | N. nova | |
| 00-025-0538 | N. nova | N. nova | N. nova | |
| 00-056-3529 | N. nova | N. nova | N. nova | |
| 00-130-2170 | N. nova | N. nova | N. nova | |
| 00-314-1789 | N. nova | N. nova | N. nova | |
| 01-097-0996 | N. nova | N. nova | N. nova | |
| 01-290-2233 | N. nova | N. nova | N. nova | |
| 02-021-0419 | N. nova | N. nova | N. nova | |
| 02-176-3564 | N. nova | N. nova | N. nova | |
| 02-190-3153 | N. nova | N. nova | N. nova | |
| 02-199-2723 | N. nova | N. nova | N. nova | |
| 02-338-3117 | N. nova | N. nova | N. nova | |
| 02-352-3316 | N. nova | N. nova | N. nova | |
| 03-023-2333 | N. nova | N. nova | N. nova | |
| 03-148-3019 | N. nova | N. nova | N. nova | |
| 03-191-3673 | N. nova | N. nova | N. nova | |
| 03-193-1476 | N. nova | N. nova | N. nova | |
| 03-209-3294 | N. nova | N. nova | N. nova | |
| 03-311-3419 | N. nova | N. nova | N. nova | |
| 04-021-3630 | N. nova | N. nova | N. nova | |
| 04-065-3069 | N. nova | N. nova | N. nova | |
| 04-110-3287 | N. nova | N. nova | N. nova | |
| 04-147-3176 | N. nova | N. nova | N. nova | |
| 04-313-0574 | N. nova | N. nova | N. nova | |
| 05-053-4454 | N. nova | N. nova | N. nova | |
| 05-141-1366 | N. nova | N. nova | N. nova | |
| 07-123-5080 | N. nova | N. nova | N. nova | |
| 07-243-3613 | N. nova | N. nova | N. nova | |
| 07-254-3806 | N. nova | N. nova | N. nova | |
| 00-188-1453 | N. otitidiscaviarum | N. otitidiscaviarum | N. otitidiscaviarum | |
| 01-257-1336 | N. otitidiscaviarum | N. otitidiscaviarum | N. otitidiscaviarum | |
| 04-009-0080 | N. otitidiscaviarum | N. otitidiscaviarum | N. otitidiscaviarum | |
| 96-247-0894c | N. transvalensis | N. blacklockiaec | N. blacklockiae | |
| 97-114-0609 | Nocardia spp. | N. paucivorans | NA | N. paucivorans |
| 02-317-3646 | Nocardia spp. | N. nova | N. nova | |
| 04-241-1298 | Nocardia spp. | N. veterana | N. veterana | |
| 04-352-2154 | Nocardia spp. | N. veterana | N. veterana | |
| 05-200-1797 | Nocardia spp. | N. paucivorans | NA | N. paucivorans |
| 06-110-2434 | Nocardia spp. | N. paucivorans | NA | N. paucivorans |
| 06-149-4352 | Nocardia spp. | N. farcinica | N. farcinica | N. farcinica |
| 06-165-4254 | Nocardia spp. | N. veterana | N. veterana | |
| 06-205-0839 | Nocardia spp. | N. paucivorans | NA | N. paucivorans |
| 06-337-1432 | Nocardia spp. | N. farcinica | N. farcinica | N. farcinica |
| 07-278-1327 | Nocardia spp. | N. veterana | N. veterana | |
| 07-296-2401 | Nocardia spp. | N. veterana | N. veterana | |
| 07-295-3727 | Nocardia spp. | N. cyriacigeorgica | N. cyriacigeorgica | |
| 02-218-3833 | Nocardia spp. | N. brevicatena | N. brevicatena | |
NA, not available; no result was determined, as an “N. paucivorans-specific” 16S rRNA gene probe was not utilized in the RLB assay.
NS, no RLB signal. This isolate's 16S rRNA gene sequence, in comparison to the reference N. asteroides GenBank sequence (GenBank accession no. AF430019) used to design the Nast-16S probe, revealed the following nucleotide polymorphisms: bp positions 151 to153, TTC to ACA, and bp positions 166 to 168, GAG to TGT.
The 606-bp 16S rRNA gene sequence of this isolate yielded 100% similarity with that of the sequence of the type strain of N. blacklockiae (GenBank accession no. EU099366).
DNA extraction.
DNA extraction was performed as previously described (19). Briefly, Nocardia cells from BHI broth cultures in the late logarithmic phase were harvested by centrifugation at 14,000 × g for 10 min. The supernatant was removed, and the pellet was suspended in 150 μl of digestion buffer (10 mM Tris-HCl [pH 8.0], 0.45% Triton X-100, 0.45% Tween 20). Bacterial suspensions were heated for 10 min at 100°C to lyse the cells, followed by cooling at −20°C for 1 h. Cell lysates were centrifuged at 14,000 × g for 5 min to pellet the cell debris. Supernatants containing DNA were diluted in 350 μl TE buffer (5 mM Tris HCl, 0.5 mM EDTA) and centrifuged for 2 min to remove cell debris. DNA was quantitated and stored at −20°C until required.
Oligonucleotide probe and primer design. (i) Primers.
Relevant sequences spanning the Nocardia 16S and 23S rRNA genes and the intervening 16S-23S rRNA gene regions were obtained from the GenBank/EMBL/DDBJ database (http://www.ncbi.nlm.nih.gov). The following primer pairs were designed: (i) 16S-27f/16S-907r and 16S-27f/16S-606rb, modified from those described previously by Becker et al. and Rodriguez-Nava et al. (3, 24) (Table 2), which were used to amplify the Nocardia 16S rRNA genes for DNA sequencing and RLB purposes, respectively, and (ii) primers 16Sf and 23Srb, used to amplify the 16S-23S rRNA gene spacer region. The reverse primers, 16S-606rb and 23Srb, were 5′-end biotin labeled (Beijing AuGCT Biotechnology Co. Ltd., Beijing, People's Republic of China).
TABLE 2.
Primers and oligonucleotide probes used in this study
| Primer or probea | Target | GenBank accession no. | Sequence (5′-3′)b |
|---|---|---|---|
| Primers | |||
| 16S-27fc | 16S rRNA gene | AB162791 | 3-AGAGTTTTGATCMTGGCTCAGG-23 |
| 16S-606rbc | 16S rRNA gene | AB162791 | 647-GAATTCCAGTCTCCCCTG-630 |
| 16S-907rc | 16S rRNA gene | AB162791 | 896-CCGTCAATTCMTTRAGTTT-877 |
| 16Sf | 16S rRNA gene | AB162791 | 1306-GAAGTCGGAGTCGCTAGTAATCGCAGATCAGC-1331 |
| 23Srb | 23S rRNA gene | AP006618 | 1181508-GACAGCTCCCCGAGGCTTATCGCA-1181485 |
| Probes | |||
| Nabs-16S | 16S rRNA gene of Nocardia abscessus | AF218292 | 140-CTGCTGTCGCATGGCGGTGG-159 |
| Nast-16S | 16S rRNA gene of Nocardia asteroides | AF430019 | 150-CTTCGGATGCATGTCTGAGG-169 |
| Nbla-16S | 16S rRNA gene of Nocardia blacklockiae | EU099366 | 125-CAGGGAGTGCATGCTCTTTG-144 |
| Nbra-16S | 16S rRNA gene of Nocardia brasiliensis | AF430038 | 150-CTTTCAGTGCATGCTGTTGG-169 |
| Nbre-16S | 16S rRNA gene of Nocardia brevicatena | AF430030 | 156-TCGCATGGCCGGGGGTGGAAA-176 |
| Ncar-16S | 16S rRNA gene of Nocardia carnea | AF430035 | 150-CTGCAGTTGCATGACTGTGG-169 |
| Ncyr/far-16S | 16S rRNA gene of Nocardia cyriacigeorgica and Nocardia farcinica | AF430027, AF430033 | 150-CTTACATCGCATGGTGTTTG-169 |
| Nfneg-16S | 16S rRNA gene of Nocardia spp. other than N. farcinica | AF430027 | 175-AAGATTTATCGGTGCGAGATGGG 197 |
| Nnov-16S | 16S rRNA gene of Nocardia nova | AF430028 | 543-TCGATTGTGAAAACTTGCAG-562 |
| Noti-16S | 16S rRNA gene of Nocardia otitidiscaviarum | DQ659912 | 168-GTGGAAAGGTTTACTGGTGCG-188 |
| Ntha-16S | 16S rRNA gene of Nocardia thailandica | AB126874 | 129-CTTGGACTGCATGGTCTTTG-148 |
| Ntra-16S | 16S rRNA gene of Nocardia transvalensis | AF430047 | 150-CACATGTCGCATGGTGTGT-168 |
| Nvet-16S | 16S rRNA gene of Nocardia veterana | ||
| ACTMY-ITS-a | 16S-23S rRNA gene spacer region of Nocardia, Rhodococcus, Gordona, Streptomyces, and Tsukamurella | EF204471 | 294-GTGTGTTGTTTGAGAACTGCACAGT-318 |
| ACTMY-ITS-b | 16S-23S rRNA gene spacer region of Nocardia, Rhodococcus, Gordona, Streptomyces, and Tsukamurella | EF204471 | 319-GGACGCGAGCATCTTTGTTA-338 |
| Nfar-ITS-a | 16S-23S rRNA gene spacer region of N. farcinica | AF536433 | 113-ATGTGCCTCGTGTATCGGTT-132 |
| Nfar-ITS-b | 16S-23S rRNA gene spacer region of N. farcinica | AF536433 | 265-GTCCAGGGTTGAGGTTTCTT-284 |
| Npau-ITS | 16S-23S rRNA gene spacer region of N. paucivorans | AF536470 | 124-TTGCTCTCGATGGGGTGGC-142 |
Abbreviations: f, forward; r, reverse; ITS, 16S-23S rRNA gene spacer region.
Numbers represent the numbered base positions where the primer or probe sequences start or finish (commencing at point 1 of the corresponding gene GenBank sequence).
(ii) Probes.
All probes were designed to achieve maximum specificity according to previously published protocols (20) after careful alignment with archived Nocardia 16S rRNA gene and 16S-23S rRNA gene spacer sequences in the GenBank/EMBL/DDBJ database by using Clustal W (30) (BioManager; Angis). The more variable first 100- to 200-bp region (5′ end) of the 16S rRNA gene was used as the basis for the design of 16S rRNA gene species-specific probes. However, for N. nova- and N. veterana-specific probes, the 500- to 600-bp region was employed, as sequences of N. nova and N. veterana in the 100- to 200-bp region revealed substantial intraspecies heterogeneity.
Initially, 14 species-specific 16S rRNA gene probes to detect 14 major Nocardia species represented in our culture collection were designed (5, 8, 19, 24, 27). Early experiments, however, identified a number of problems. First, the “N. paucivorans-specific” 16S rRNA gene probe cross-hybridized with several other species including N. nova and was not further utilized. Second, the “N. farcinica-specific” probe failed to hybridize to a proportion of N. farcinica isolates including the ATCC strains due to the substantial intraspecies genetic heterogeneity in the region used for the probe design. It also cross-hybridized with several uncommon Nocardia species and likewise was not further used. However, we identified a nucleotide region (bp positions ∼150 to 200) common to all species in the test set of isolates other than N. farcinica and designed a group-specific probe, Nfneg-16S (Table 2), to detect these isolates; initial testing showed that N. farcinica isolates produced no RLB signal with this probe. Finally, the design of an “N. cyriacigeorgica-specific” probe was precluded by intraspecies heterogeneity and an inability to distinguish between this species and N. farcinica. Instead, the dual-species Ncyr/far-16S probe (Table 2) was designed to detect N. farcinica and N. cyriacigeorgica.
Two N. farcinica-specific probes and one N. paucivorans-specific probe targeting the 16S-23S rRNA gene spacer region were also designed (Table 2); these probes were used to identify isolates that could not be identified to the species level by the 16S rRNA gene probes (see Results). Additionally, two taxon-specific 16S-23S rRNA gene spacer-directed probes (ACTMY-ITS-a and ACTMY-ITS-b) (Table 2) were incorporated into the RLB assay in order to “capture” as many known Nocardia spp. as possible, which can then be classified by use of the other probes. These probes may hybridize with DNA from other species of aerobic actinomycetes. We were unable to design genus-specific ITS probes targeted only at Nocardia spp.; the successful design of such probes may require complete sequence data for multiple species of Nocardia.
Thus, a total of 18 probes were employed in the RLB assay: 11 16S rRNA gene-targeted species-specific probes, two group-specific 16S rRNA gene probes, and five 16S-23S rRNA gene spacer-targeted probes (Table 2). All probes were 5′-hexylamine labeled and synthesized commercially (Beijing AuGCT Biotechnology Co. Ltd.).
PCR in preparation for RLB assays.
The 16S rRNA gene region was amplified by using primer pair 16S-27f/16S-606rb, while the 16S-23S rRNA gene spacer region was amplified with primer pair 16Sf/23Srb (Table 2). Each PCR mixture contained 5 μl template DNA, 3 μl of 10× PCR buffer (Qiagen, Doncaster, Victoria, Australia), 0.2 mM each deoxynucleoside triphosphate (dNTP) (Roche Diagnostics, Mannheim, Germany), 1.5 U HotStar Taq polymerase (Qiagen), 0.25 μl (50 pmol/μl) each forward primer and reverse primer, and water to a final volume of 30 μl. Amplification was performed by use of a Mastercycler gradient thermocycler (Eppendorf GmbH, Germany). The cycling conditions were 95°C for 15 min followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 90 s, with a final extension step at 72°C for 10 min. PCR products (6 to 8 μl) were visualized on a 2% agarose gel after staining with SYBR safe DNA gel stain (Invitrogen, Mt. Waverley, Victoria, Australia).
RLB hybridization assay.
The RLB assay was performed as described previously (20). Briefly, the probes (final concentration of approximately 10 μM in 0.5 M NaHCO3 [pH 8.4]) were covalently linked to a Biodyne C nylon membrane (Pall Life Sciences, Ann Arbor, MI), which was “negatively activated” by incubation in 16% (wt/vol) 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDAC) (Sigma, St. Louis, MO). The membrane was then placed into an MN45 miniblotter (Immunetics, Boston, MA). The amplified PCR product (20 μl) was diluted into 150 μl of 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])-0.1% sodium dodecyl sulfate (SDS), denatured at 100°C for 10 min, and cooled immediately on ice.
Hybridization was performed by adding 150 μl of diluted denatured PCR product to membrane-bound probes at 60°C for 1 h. The membrane was washed twice (10 min each time) at 60°C with 2× SSPE-0.5% SDS and incubated in peroxidase-labeled streptavidin conjugate (Roche) at 42°C for 1 h. The membrane was further washed with 2× SSPE buffer-0.5% SDS at 42°C and then at 25°C. Bound PCR products were detected by chemiluminescence by using ECL detection liquid (Amersham, GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) and visualized by exposure to a Hyperfilm ECL X-ray film (Amersham) for 6 min.
16S rRNA gene sequence analysis.
16S rRNA gene sequencing was performed for comparison with results obtained by RLB analysis; for isolates received since 2005, this was performed routinely as a diagnostic service. The 5′ end of the 16S rRNA gene region was amplified by using primer pair 16S-27f/16S-907r (as described above). PCR products were purified by using a PCR product presequencing kit (USB Corporation, Cleveland, OH) and sequenced by using forward (16S-27f) and reverse (16S-907r) primers (Table 2) and the BigDye Terminator (version 3.1) cycle sequencing kit (ABI Prism 3100; Applied Biosystems, Foster City, CA). Sequences were aligned by using BLASTn 2.2.10 (2), accessed via BioManager. A similarity score of ≥99% between the unknown sequence and the reference database sequence(s) was used as the criterion to classify an isolate to the species level (7, 25), while a similarity score of 97 to 98.9% identified an isolate as belonging to the genus Nocardia but belonging to a different species (6, 18).
RESULTS
The modified universal bacterial primers 16S-27f and 16S-606rb amplified the 16S rRNA gene region, and primer pair 16Sf/23Srb amplified the 16S-23S rRNA gene spacer region for all test isolates. Species identification of ATCC isolates was determined by PCR/RLB analysis and 16S rRNA gene sequencing.16S rRNA gene-targeted probes (see Materials and Methods) provided the correct identification of 11 of the 12 (92%) strains including N. carnea and N. veterana but were unable to assign N. paucivorans ATCC BAA-278T to its species. However, the N. paucivorans-specific 16S-23S rRNA gene spacer probe provided a clear RLB signal when tested on N. paucivorans ATCC BAA-278T. Concordant results with DNA sequencing were obtained for all isolates with both probe sets.
Clinical isolates.
Species identification determined by phenotype-based methods, PCR/RLB analysis, and 16S rRNA gene sequencing of clinical isolates is summarized in Table 1.
(i) Sensitivity of 16S rRNA gene-targeted RLB assay.
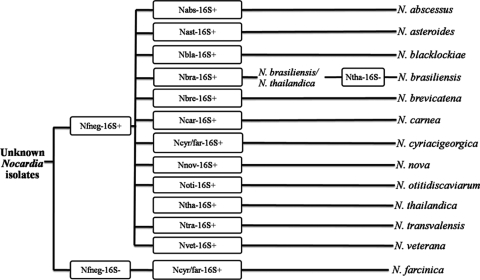
The 16S rRNA gene-targeted RLB assay correctly identified 113 of 123 clinical isolates to the species level (sensitivity, 91.8%), including uncommon and novel Nocardia species and 10 isolates identified only to the genus level using phenotypic methods (Table 1). All N. farcinica strains were “flagged” as such by the absence of an RLB signal with probe Nfneg-16S and then identified with subsequent hybridization with probe Ncyr/Nfar-16S (Fig. 1 and 2). Isolates were assigned as being N. cyriacigeorgica isolates where hybridization occurred with the Nfneg-16S and Ncyr/Nfar-16S probes (Fig. 1).
FIG. 1.
Algorithm for identification of a suspected Nocardia sp. (also positive signal with the ACTMY-ITS-a and ACTMY-ITS-b probes) (Table 2) using 11 Nocardia species-specific and two group-specific probes targeted at the 16S rRNA gene region in an RLB assay.
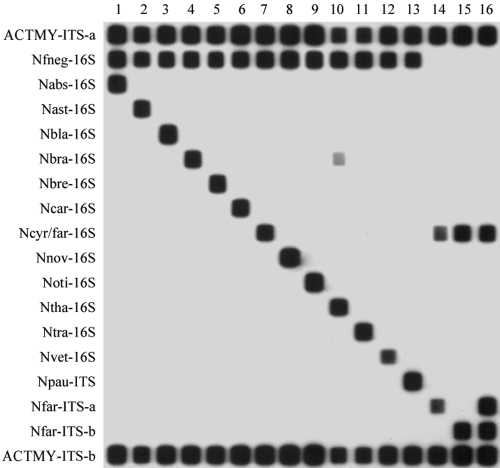
FIG. 2.
RLB results for 16 Nocardia isolates representing 14 species. Lane 1, isolate ATCC BAA-279T (N. abscessus); lane 2, isolate ATCC 19247T (N. asteroides); lane 3, isolate 96-247-0894 (N. blacklockiae); lane 4, isolate ATCC 19296 (N. brasiliensis); lane 5, isolate ATCC 15333T (N. brevicatena); lane 6, isolate ATCC 6847 (N. carnea); lane 7, isolate 00-159-1584 (N. cyriacigeorgica); lane 8, isolate ATCC 33726T (N. nova); lane 9, isolate ATCC 14629T (N. otitidiscaviarum); lane 10, isolate 00-355-2649 (N. thailandica); lane 11, ATCC 6865T (N. transvalensis); lane 12, isolate ATCC BAA-509 (N. veterana); lane 13, isolate ATCC BAA-278T (N. paucivorans); lanes 14 to 16, isolates ATCC 3308, 05-048-3600, and 07-023-0418 (all N. farcinica isolates). The left column shows the probe list. The blot shows (i) the taxon-specific probes ACTMY-ITS-a and ACTMY-ITS-b to flag all isolates as possible Nocardia spp., (ii) the group-specific probe Nfneg-16S hybridizing with all Nocardia species tested other than N. farcinica, (iii) the group-specific probe Ncyr/far-16S hybridizing with both N. cyriacigeorgica and N. farcinica, (iv) weak cross-reaction between probe Nbra-16S and DNA from N. thailandica, and (v) different N. farcinica isolates hybridizing with either or both of the Nfar-ITS-a and Nfar-ITS-b probes.
Of the 10 isolates that were unable to be identified with the 16S-taregted probes, no RLB signal was obtained for one strain (isolate 02-071-3627). This isolate was phenotypically classified as belonging to the N. asteroides complex and confirmed as being N. asteroides sensu stricto by 16S rRNA gene sequence analysis (Table 1). Examination of its sequence in comparison with the reference N. asteroides GenBank sequence (GenBank accession no. AF430019) used to design the Nast-16S probe (Table 2) revealed a number of nucleotide polymorphisms in the region employed for probe design, bp positions 151 to 153, TTC to ACA, and bp positions 166 to 168, GAG to TGT; furthermore, this Nocardia sequence has not been reported previously. The other two N. asteroides sensu stricto isolates tested (strains ATCC 19247T and 04-128-3054) (Table 1) produced a clear RLB signal with the Nast-16S probe. The remaining nine isolates not assigned to the species level were classified as being N. paucivorans isolates by the N. paucivorans-specific 16S-23S rRNA gene spacer (Npau-ITS) probe and by DNA sequencing (Table 1 and see below).
(ii) Specificity of 16S rRNA gene-targeted RLB assay.
For clinical isolates, 16S rRNA gene species-specific probes correctly identified isolates of 11 major Nocardia species, N. abscessus, N. asteroides, Nocardia blacklockiae, N. brasiliensis, Nocardia brevicatena, N. cyriacigeorgica, N. nova, N. otitidiscaviarum, Nocardia thailandica, and N. veterana, with no cross-reactivity among the species studied with only one exception. The Nbra-16S probe produced a weak hybridization signal with N. thailandica; however, the latter can be identified clearly by hybridization with the highly specific Ntha-16S probe (Table 2 and Fig. 1). The dual-species Ncyr/far-16S probe hybridized only with N. cyriacigeorgica and N. farcinica. This probe was able to definitively identify those isolates that failed to hybridize with the Nfneg-16S probe (and, by default, are provisionally assigned as “N. farcinica”) as N. farcinica isolates (Fig. 2).
16S-23S rRNA gene spacer (ITS) probes.
The taxon-specific ACTMY-ITS-a and ACTMY-ITS-b probes (Table 2) hybridized with amplified DNA of all study isolates. Of note, the N. paucivorans-specific probe (Npau-ITS) correctly identified the type strain (strain ATCC BAA-278T) and all nine clinical N. paucivorans isolates with high specificity (see above). There was no cross-hybridization with DNA of other Nocardia species. Thirteen of 21 (62%) N. farcinica (19 clinical and 2 ATCC strains) isolates produced a hybridization signal with both the Nfar-ITS-a and Nfar-ITS-b probes. Four isolates (17%) hybridized only with the Nfar-ITS-a probe, while an RLB signal with the Nfar-ITS-b probe only was noted for the remaining four strains; the use of either probe alone correctly identified 17 (81%) strains, but the application of both probes led to the correct identification of all 21 strains.
Thus, the combination of 16S rRNA gene and 16S-23S rRNA gene spacer-targeted probes identified 122 of 123 (99.2%) isolates (13 species) to the species level. The concordance with 16S rRNA gene sequencing was 100% for those strains (n = 122) for which a hybridization signal was obtained.
Proposed set of probes for RLB-based identification of Nocardia spp.
A proposed set of probes for incorporation into a PCR/RLB assay for the identification of 14 Nocardia species is shown in Fig. 2, while a guide to the species identification of Nocardia isolates based on the results obtained with the 16S rRNA gene probes only is given in Fig. 1. The Nfneg-16S probe potentially separates N. farcinica from other major Nocardia spp. and may be used in parallel with each of the 11 16S rRNA gene species-specific and Ncyr/far16S probes for species identification (Fig. 1 and 2). The incorporation of the Npau-ITS probe is required to identify N. paucivorans, while the two 16S-23S rRNA gene spacer N. farcinica-specific probes increase the sensitivity of the assay. Because it cannot be excluded that closely related species not targeted by the RLB assay may cross-hybridize with a particular species-specific probe (e.g., the uncommon species Nocardia arthritidis is expected to produce an RLB signal with the N. abscessus-specific 16S rRNA gene probe due to identical 16S rRNA gene sequences in the region of probe design), additional examination of the isolate may be required (see Discussion).
Species distribution of clinical isolates.
By PCR/RLB and 16S rRNA gene sequencing, 12 Nocardia species among 123 clinical isolates were clearly identified, with the most common identification being N. cyriacigeorgica (34 isolates; 27.6%), followed by N. nova (33 isolates; 26.8%), N. farcinica (n = 19; 15.4%), and N. paucivorans (n = 9; 7.3%). There were eight isolates each of N. brasiliensis and N. veterana, four isolates of N. otitidiscaviarum, three strains of N. abscessus, two of N. asteroides, and one each of N. brevicatena, N. blacklockiae, and N. thailandica (Table 1).
DISCUSSION
The molecular-based identification of Nocardia spp. remains a challenge with the increasing recognition of novel species and taxonomic reassignment (5, 23, 25, 27). The present study demonstrates the potential of combining a novel broad-range Nocardia PCR with probe hybridization technology in an RLB format to identify clinically relevant Nocardia species including newly described species. Features of the RLB assay, which incorporates a panel of 16S as well as 16S-23S rRNA gene spacer-targeted probes, include a high sensitivity (overall, >99%) and specificity for the identification of the 14 Nocardia species studied.
The validity of the RLB assay was confirmed by the study of a large number of reference and clinical Nocardia strains in comparison with 16S rRNA gene sequencing data. Although the 16S rRNA gene is highly conserved, the first 500 bp of the gene is well known to contain variable regions that enable species discrimination (reviewed in reference 5). Our results indicate that overall, this region can also be successfully targeted in probe-based identification strategies. However, two problems hampered efficient probe design. First, we were unable to identify signature polymorphisms in the region used for probe design to distinguish (i) N. paucivorans from other species and (ii) N. farcinica from N. cyriacigeorgica. The explanation for this is not readily apparent, but it is possible that probes directed at identifying these species should be designed based on less conserved regions of the Nocardia genome. Second, we encountered substantial intraspecies sequence variation within N. farcinica and N. cyriacigeorgica, which precluded the design of reliable species-specific 16S-targeted probes for these species. It is likely that short (20- to 30-bp) single probes are sufficiently sensitive to identify only some, but not all, isolates of these species (data not shown). Intraspecies sequence heterogeneity was also observed for N. nova, but in this case, the Nnov-16S probe hybridized with all N. nova isolates (Table 1).
The 16S rRNA gene-targeted probes demonstrated good sensitivity (91.8%) among clinical isolates. The inability of the assay to identify a single N. asteroides sensu stricto isolate (Table 1) can be explained by the presence of nucleotide polymorphisms in the region of probe design compared with the reference 16S rRNA gene sequence of N. asteroides (GenBank accession no. AF430019) employed; indeed, the sequence of this isolate was a novel sequence. Furthermore, N. asteroides sensu stricto (as determined by gene sequencing) is now considered a rare pathogen (5). The RLB assay was also unable to detect and/or identify N. paucivorans isolates (see below). The identification of N. farcinica and N. cyriacigeorgica strains was achieved by observing results of hybridization with two “group-specific” probes (Nfneg-16S and Ncyr/Nfar-16S) (Fig. 1).
The RLB assay was specific for the test set of isolates, with one exception: the weak cross-reactivity between the N. brasiliensis-specific 16S probe and N. thailandica DNA is unexplained (Fig. 1). It is possible, however, that certain species not targeted by the RLB assay may cross-hybridize with target species-specific probes. For instance, the uncommon pathogens N. arthritidis and N. asiatica are expected to produce an RLB signal with the Nabs-16S probe (Table 2), since these species share indistinguishable 16S rRNA gene sequences in the region used for probe design; we were unable to test this prediction since isolates of either species were not available for study. Nonetheless, 16S rRNA gene sequence polymorphisms were able to clearly distinguish between closely related species, most notably N. nova and N. veterana, two species with 98.1% 16S rRNA gene sequence similarity (15, 25). Furthermore, sequence polymorphisms in the region of probe design based on GenBank sequence alignments (Table 2) are predicted to enable the differentiation of the closely related Nocardia elegans and Nocardia africana from N. nova as well as from N. veterana. Isolates of N. elegans and N. africana likewise were not available for analysis.
To “capture” as many Nocardia species as possible that are not targeted in the assay, we incorporated two taxon-specific (ACTMY-ITS-a and ACTMY-ITS-b) probes into the RLB format; although the probes may also hybridize with DNA from other aerobic actinomycetes, these organisms are usually distinguished from Nocardia species by phenotypic methods (13, 21). We chose to target nucleotide polymorphisms in the 16S-23S rRNA gene spacer region since this region is less conserved than the 16S locus. Cross-hybridization of other actinomycete species with Nocardia species- or group-specific probes was not observed, with the exception of a small number (<5) of Rhodococcus equi strains with the N. abscessus-specific and N. otitidiscaviarum-specific 16S rRNA gene probes (data not shown). However, in every instance, phenotypic methods were sufficient to identify the organism as being R. equi, so they would not have been subjected to analysis by PCR/RLB.
Another key finding of the present study includes the recognition of nucleotide polymorphisms within the 16S-23S rRNA gene spacer locus as a potentially useful target for the identification of Nocardia spp. Our results further support the general notion of the use of more than one genetic locus for species determination (5, 12, 24). Indeed, the incorporation of probes targeted at the 16S-23S rRNA gene spacer region into the RLB assay as an additional, although smaller, set of probes allowed the 14 test Nocardia species to be identified with an increased sensitivity (99.2%) and high specificity (99%). In particular, the N. paucivorans-specific 16S-23S rRNA gene spacer probe enabled the species identification of N. paucivorans, which was not achievable with the 16S rRNA gene-based probe set (as described above). The 16S-23S rRNA gene spacer locus was also more discriminatory for the identification of N. farcinica (Fig. 2). The concordance with 16S rRNA gene sequencing was 100% for all isolates for which the RLB assay assigned species.
The results also indicate that the RLB assay has the potential to identify subtypes or genotypes of Nocardia isolates since the two N. farcinica-specific 16S-23S rRNA gene spacer probes hybridized with different (as well as with the same) isolates. This likely reflects intraspecies sequence variation within the 16S-23S rRNA gene spacer region for N. farcinica, necessitating the inclusion of >1 probe to detect N. farcinica to optimize sensitivity. Further studies examining subtypes of N. farcinica or of other species in the context of epidemiological or disease associations are warranted given that sequence (16S rRNA gene)-based subtypes have recently been described for some species including N. nova and N. cyriacigeorgica (19, 27).
Because of the diverse range of potential nocardial pathogens and the importance of species identification for clinical decisions, 16S rRNA gene sequencing remains the preferred method for definitive species determination. We confirm its accuracy in this capacity (this study; 5). However, this approach is expensive in Australia (AU$15/US$14 per isolate) and is impractical for analyses of large numbers of strains. Multiplex PCR assays are alternative identification strategies but are limited by the number of primers that are able to be incorporated. Probe-based hybridization technologies for Nocardia identification are not well established, although one real-time platform was evaluated with a limited number of strains (1). We chose the RLB format due to its relative simplicity and ability to simultaneously analyze multiple isolates against multiple probes. By employing 16S rRNA gene as well as 16S-23S rRNA gene spacer-targeted probes, the assay demonstrated a sensitivity comparable to that of 16S rRNA gene sequencing. However, in its current design, laboratories may also consider 16S rRNA gene sequencing to provide a 2-step (by RLB assay and sequencing) confirmation process for isolates assigned as being (i) N. cyriacigeorgica by the RLB assay, as the identification of this species is inferred from the hybridization pattern using two group-specific probes (Fig. 1), and (ii) N. abscessus, since N. arthritidis and N. asiatica may produce an RLB signal with the Nabs-16S probe; the latter two species are rare pathogens. Nevertheless, the RLB assay described here is a useful adjunct to DNA sequencing for the species identification of Nocardia isolates, especially if there is a need for the simultaneous identification of >1 isolate. Although only 18 probes were utilized in the present study, up to 40 may be incorporated, extending the range of pathogens that can be identified. This flexibility allows laboratories to customize the RLB format to meet requirements for the inclusion of target species according to local epidemiology.
An additional limitation of the present assay (and of other molecular assays) is time. A full working day is required for DNA extraction, PCR (3 h), and product detection and identification using the RLB assay (5 h). However, the RLB membrane can be reused at least 20 times, and running costs associated with the assay are less (AUS$8/US$7 per isolate) than those for DNA sequencing. We will now evaluate this assay by broadening the range of target species and also by determining its ability to detect Nocardia isolates in clinical specimens including tissue, bronchoalveolar lavage fluid, and whole blood.
In conclusion, this study presents for the first time a combined PCR/RLB assay for the species identification of Nocardia isolates and the algorithm for its use in a diagnostic laboratory. In the event that a suspected “Nocardia” isolate is not identified further due to a lack of inclusion of a species-specific probe or where species assignment is ambiguous, 16S rRNA gene sequencing should be performed. An evaluation of larger numbers of isolates/species and the testing of its capability to directly detect Nocardia isolates in clinical specimens are required to position the RLB assay within routine diagnostic algorithms.
ADDENDUM
Following the completion of the study and after the time of writing, we acquired a single N. elegans strain. This isolate did not hybridize with any of the Nocardia probes including the N. veterana- and N. nova-specific 16S rRNA gene-targeted probes.
Acknowledgments
We thank Catriona Halliday for assistance with isolate identification, Maryann Pincevic for her assistance in performing the 16S rRNA gene sequencing, and Ping Zhu for help with the figures.
Footnotes
Published ahead of print on 2 December 2009.
REFERENCES
- 1.Alfaresi, M., and A. Elkosh. 2006. Rapid identification of clinically relevant Nocardia species using real-time PCR with SYBR green and melting-curve analysis. J. Med. Microbiol. 55:1711-1715. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, K., D. Harmsen, A. Mellmann, C. Meier, P. Schumann, G. Peters, and C. von Eiff. 2004. Development and evaluation of a quality-controlled ribosomal sequence database for 16S ribosomal DNA-based identification of Staphylococcus species. J. Clin. Microbiol. 42:4988-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, J. M., K. N. Pham, M. M. McNeil, and B. A. Lasker. 2004. Rapid identification of Nocardia farcinica clinical isolates by a PCR assay targeting a 314-base-pair species-specific DNA fragment. J. Clin. Microbiol. 42:3655-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown-Elliott, B. A., J. M. Brown, P. S. Conville, and R. J. Wallace, Jr. 2006. Clinical and laboratory features of the Nocardia spp. based on current molecular taxonomy. Clin. Microbiol. Rev. 19:259-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarridge, J. E., III. 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17:840-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cloud, J. L., P. S. Conville, A. Croft, D. Harmsen, F. G. Witebsky, and K. C. Carroll. 2004. Evaluation of partial 16S ribosomal DNA sequencing for identification of Nocardia species by using the MicroSeq 500 system with an expanded database. J. Clin. Microbiol. 42:578-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conville, P. S., J. M. Brown, A. G. Steigerwalt, B. A. Brown-Elliott, and F. G. Witebsky. 2008. Nocardia wallacei sp. nov. and Nocardia blacklockiae sp. nov., human pathogens and members of the “Nocardia transvalensis complex.” J. Clin. Microbiol. 46:1178-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conville, P. S., and F. G. Witebsky. 2004. Current issues pertaining to the Nocardia species. Clin. Microbiol. Newsl. 26:57-62. [Google Scholar]
- 10.Conville, P. S., and F. G. Witebsky. 2005. Multiple copies of the 16S rRNA gene in Nocardia nova isolates and implications for sequence-based identification procedures. J. Clin. Microbiol. 43:2881-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conville, P. S., and F. G. Witebsky. 2007. Analysis of multiple differing copies of the 16S rRNA gene in five clinical isolates and three type strains of Nocardia species and implications for species assignment. J. Clin. Microbiol. 45:1146-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conville, P. S., A. M. Zelazny, and F. G. Witebsky. 2006. Analysis of secA1 gene sequences for identification of Nocardia species. J. Clin. Microbiol. 44:2760-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conville, P. S., and F. G. Witebsky. 2007. Nocardia, Rhodococcus, Gordonia, Actinomadura, Streptomyces, and other aerobic actinomycetes, p. 515-542. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
- 14.Gold, B. 2003. Origin and utility of the reverse dot-blot. Expert Rev. Mol. Diagn. 3:143-152. [DOI] [PubMed] [Google Scholar]
- 15.Gurtler, V., R. Smith, B. C. Mayall, G. Potter-Reinemann, E. Stackebrandt, and R. M. Kroppenstedt. 2001. Nocardia veterana sp. nov., isolated from human bronchial lavage. Int. J. Syst. Evol. Microbiol. 51:933-936. [DOI] [PubMed] [Google Scholar]
- 16.Hasegawa, T., T. Gonoi, J. Ito, T. Kogure, K. Yazawa, and Y. Mikami. 2007. Identification of Nocardia farcinica by a PCR primer amplifying a specific DNA band for the bacterium. Nippon Ishinkin Gakkai Zasshi 48:173-175. (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 17.Isik, K., and M. Goodfellow. 2002. Differentiation of Nocardia species by PCR-randomly amplified polymorphic DNA fingerprinting. Syst. Appl. Microbiol. 25:60-67. [DOI] [PubMed] [Google Scholar]
- 18.Janda, J. M., and S. L. Abbott. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45:2761-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong, F., S. C. Chen, X. Chen, V. Sintchenko, C. Halliday, L. Cai, Z. Tong, O. C. Lee, and T. C. Sorrell. 2009. Assignment of reference 5′-end 16S rDNA sequences and species-specific sequence polymorphisms improves species identification of Nocardia. Open Microbiol. J. 3:97-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong, F., and G. L. Gilbert. 2006. Multiplex PCR-based reverse line blot hybridization assay (mPCR/RLB)—a practical epidemiological and diagnostic tool. Nat. Protoc. 1:2668-2680. [DOI] [PubMed] [Google Scholar]
- 21.McNeil, M. M., and J. M. Brown. 1994. The medically important aerobic actinomycetes: epidemiology and microbiology. Clin. Microbiol. Rev. 7:357-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minero, M. V., M. Marin, E. Cercenado, P. M. Rabadan, E. Bouza, and P. Munoz. 2009. Nocardiosis at the turn of the century. Medicine 88:250-261. [DOI] [PubMed] [Google Scholar]
- 23.Patel, J. B., R. J. Wallace, Jr., B. A. Brown-Elliott, T. Taylor, C. Imperatrice, D. G. Leonard, R. W. Wilson, L. Mann, K. C. Jost, and I. Nachamkin. 2004. Sequence-based identification of aerobic actinomycetes. J. Clin. Microbiol. 42:2530-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez-Nava, V., A. Couble, G. Devulder, J. P. Flandrois, P. Boiron, and F. Laurent. 2006. Use of PCR-restriction enzyme pattern analysis and sequencing database for hsp65 gene-based identification of Nocardia species. J. Clin. Microbiol. 44:536-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roth, A., S. Andrees, R. M. Kroppenstedt, D. Harmsen, and H. Mauch. 2003. Phylogeny of the genus Nocardia based on reassessed 16S rRNA gene sequences reveals underspeciation and division of strains classified as Nocardia asteroides into three established species and two unnamed taxons. J. Clin. Microbiol. 41:851-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saubolle, M. A., and D. Sussland. 2003. Nocardiosis: review of clinical and laboratory experience. J. Clin. Microbiol. 41:4497-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlaberg, R., R. C. Huard, and P. Della-Latta. 2008. Nocardia cyriacigeorgica, an emerging pathogen in the United States. J. Clin. Microbiol. 46:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steingrube, V. A., B. A. Brown, J. L. Gibson, R. W. Wilson, J. Brown, Z. Blacklock, K. Jost, S. Locke, R. F. Ulrich, and R. J. Wallace, Jr. 1995. DNA amplification and restriction endonuclease analysis for differentiation of 12 species and taxa of Nocardia, including recognition of four new taxa within the Nocardia asteroides complex. J. Clin. Microbiol. 33:3096-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su, S. C., M. Vaneechoutte, L. Dijkshoorn, Y. F. Wei, Y. L. Chen, and T. C. Chang. 2009. Identification of non-fermenting Gram-negative bacteria of clinical importance by an oligonucleotide array. J. Med. Microbiol. 58:596-605. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallace, R. J., Jr., M. Tsukamura, B. A. Brown, J. Brown, V. A. Steingrube, Y. S. Zhang, and D. R. Nash. 1990. Cefotaxime-resistant Nocardia asteroides strains are isolates of the controversial species Nocardia farcinica. J. Clin. Microbiol. 28:2726-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, Y., F. Kong, Y. Yang, and G. L. Gilbert. 2008. A multiplex PCR-based reverse line blot hybridization (mPCR/RLB) assay for detection of bacterial respiratory pathogens in children with pneumonia. Pediatr. Pulmonol. 43:150-159. [DOI] [PubMed] [Google Scholar]
- 33.Xiong, L., F. Kong, Y. Yang, J. Cheng, and G. L. Gilbert. 2006. Use of PCR and reverse line blot hybridization macroarray based on 16S-23S rRNA gene internal transcribed spacer sequences for rapid identification of 34 Mycobacterium species. J. Clin. Microbiol. 44:3544-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng, X., F. Kong, C. Halliday, S. Chen, A. Lau, G. Playford, and T. C. Sorrell. 2007. Reverse line blot hybridization assay for identification of medically important fungi from culture and clinical specimens. J. Clin. Microbiol. 45:2872-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]