Table 1. Data-processing, refinement and model statistics.
Values in parentheses are for the highest resolution shell.
| Crystal data | |
| Space group | P21 |
| Z | 4 |
| Unit-cell parameters (Å, °) | a = 30.73, b = 74.85, c = 50.52, β = 107.80 |
| Data-processing statistics | |
| Resolution (Å) | 29.54–1.60 (1.66–1.60) |
| No. of unique reflections | 26265 |
| Redundancy | 2.61 (2.15) |
| 〈I/σ(I)〉 | 16.8 (2.4) |
| Completeness | 91.2 (64.2) |
| Rmerge† | 0.027 (0.340) |
| Structure-refinement statistics | |
| Resolution (Å) | 29.54–1.60 (1.64–1.60) |
| No. of reflections (Fo = 0 removed) | 25900 (2256) |
| R/Rfree‡ (all data) | 0.197/0.253 (0.356/0.411) |
| Reflections in test set (%) | 9.80 (5.76) |
| No. of refined parameters | 8938 |
| No. of reflections | 23360 |
| No. of restraints | 14567 |
| Data-to-parameter ratio | 2.61 |
| Data/restraints-to-parameter ratio | 4.24 |
| Model statistics | |
| Non-H atoms | |
| Protein atoms (full/partial) | 1871/62 |
| Ligand atoms (full/partial) | 37/10 |
| Water atoms (full/partial) | 181/42 |
| Geometry: r.m.s. deviations from ideal values | |
| Bonds (Å) | 0.011 |
| Angles (°) | 1.55 |
| Planes (Å) | 0.010 |
| Chiral centers (Å3) | 0.09 |
| Average isotropic B factors (Å2) | |
| B estimate (Wilson) | 33.2 |
| Overall | 38.7 |
| Protein (full/partial) | 38.1/34.4 |
| Ligands (all) | 48.1/47.2 |
| Waters (full/partial) | 43.6/37.2 |
| Ramachandran plot | |
| Most favored region | 199 [86.5%] |
| Allowed region | 31 [13.5%] |
| Generously allowed and disallowed regions | 0 [0.0%] |
R
merge = 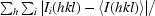
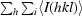 , where I
i(hkl) is the ith used observation for unique hkl and 〈I(hkl)〉 is the mean intensity for unique hkl.
, where I
i(hkl) is the ith used observation for unique hkl and 〈I(hkl)〉 is the mean intensity for unique hkl.
R = 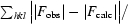
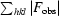 , where F
obs and F
calc are the observed and calculated structure-factor amplitudes.
, where F
obs and F
calc are the observed and calculated structure-factor amplitudes.
