The expression, purification and crystallization of the collagen-binding region of the E. rhusiopathiae surface protein RspB is described. The crystals diffracted to 2.2 Å resolution using synchrotron radiation.
Keywords: RspB, Erysipelothrix rhusiopathiae, collagen binding
Abstract
RspB is a surface adhesin of Erysipelothrix rhusiopathiae. A recombinant form of the collagen-binding region of this protein, RspB(31–348), has been overexpressed in Escherichia coli in native and selenomethionine-derivative forms and purified using affinity and gel-permeation chromatography. Thin plate-like crystals were obtained by the hanging-drop vapour-diffusion method using the same condition for both forms. The native crystals diffracted to a resolution of 2.5 Å using an in-house X-ray source, while the selenomethionine-derivative crystals diffracted to a resolution of 2.2 Å using synchrotron radiation. The crystals belonged to the monoclinic space group P21, with unit-cell parameters a = 46.19, b = 66.65, c = 101.72 Å, β = 94.11°.
1. Introduction
The proteins displayed on the surface of most Gram-positive bacteria exhibit adhesive properties and either behave as ligands to the receptors of the target cells or have specific affinity for host components such as immunoglobulins, plasma proteins or ECM (extracellular matrix) such as collagen, fibronectin, laminin etc. These surface proteins therefore colonize a particular ecological niche, the primary step in pathogenesis, and hence play an important role in virulence (Navarre & Schneewind, 1999 ▶; Scott & Barnett, 2006 ▶). Bacterial infection can often be treated successfully using antibiotics. However, antibiotic resistance is becoming more and more of a problem and even simple infections are becoming hard to treat. Therefore, new therapeutic strategies to prevent and treat infections are absolutely necessary. One such strategy is the blocking or inhibition of the primary stages of infection, namely bacterial adherence to host tissues. Since bacterial surface proteins establish the first contact with host tissues, their adherence mechanism is a critical step in successful infection. To execute the strategy of interfering with the adherence mechanism, a detailed molecular analysis of surface proteins and their interactions with host components is necessary.
Erysipelothrix rhusiopathiae is a slender Gram-positive rod which causes erysipelas in animals and erysipeloid in humans (Wood, 1999 ▶). It was first isolated by Koch in 1876 and its name literally means ‘erysipelas thread of red disease’. This pathogen infects a large variety of animals from house flies to wild bears. Infections have been reported worldwide in as many as 50 different types of animals (Gorby & Peacock, 1998 ▶), with the highest occurrence being found in domestic swine (Nerad & Snydman, 1998 ▶). E. rhusiopathiae also causes infections in humans. There are three clinical categories for the human disease caused by this organism: a localized cutaneous form (most common), a generalized cutaneous form and a septicaemic form (associated with the heart disease endocarditis). In humans, infection is primarily associated with occupational exposure: butchers, slaughterhouse workers, veterinarians, farmers and fish handlers have the highest risk of acquiring infection (Norman & Kihlstrom, 1985 ▶; Robson et al., 1998 ▶; Venditti et al., 1990 ▶).
E. rhusiopathiae primarily adheres firmly to the host tissue with the help of its surface adhesins, establishes an ecological niche which is able to resist any sort of flushing by the host and enters via scratches or puncture wounds on the surface of the skin. To date, two surface adhesins, RspA and RspB (rhusiopathiae surface protein A and rhusiopathiae surface protein B), with molecular masses of 219 and 85 kDa, respectively, have been characterized from E. rhusiopathiae. They showed structural similarity to many surface-adhesive proteins of Gram-positive bacteria (Shimoji et al., 2003 ▶). The Rsp proteins, which have a low degree of sequence identity to each other, not only bind to ECM molecules such as collagen and fibrinogen but also to abiotic (polystyrene) surfaces. They therefore constitute a novel class of cell-surface components which are involved in the initial step of biofilm formation (Shimoji et al., 2003 ▶). Biofilm formation hinders antibiotic penetration and impairs the host defence system, thereby rendering virulence to the bacteria.
The N-terminal regions of Rsp proteins showed characteristics of a collagen-binding domain similar to those described for Staphylococcus aureus Cna and Enterococcus faecalis Ace (Shimoji et al., 2003 ▶). Structurally, the 787-amino-acid residue protein (85 kDa) RspB consists of (i) an N-terminal ligand-binding region preceded by a signal peptide for secretion, (ii) a repeat domain of unknown function and (iii) a C-terminal domain containing the LPXTG motif which is followed by a hydrophobic region and positively charged amino acids which are necessary for translocation and anchoring to the cell wall (Fig. 1 ▶). Comparison of the collagen-binding region of RspB (residues 31–348) with the corresponding regions of S. aureus Cna (Rich et al., 1998 ▶) and En. faecalis Ace (Rich et al., 1999 ▶) showed a sequence identity of 27% and 25%, respectively. Here, we report a preliminary structural analysis of the collagen-binding region of RspB.
Figure 1.
Structural organization of E. rhusiopathiae RspB. The signal peptide, cell-wall anchoring domain, membrane-spanning domain and positively charged C-terminal domain are indicated by S, W, M and C, respectively.
2. Materials and methods
2.1. Cloning and expression
The rspB gene encoding the region 31–348 (devoid of the signal peptide) was PCR-amplified from the genomic DNA of E. rhusiopathiae Fujisawa and cloned into expression vector pQE30 as previously reported (Shimoji et al., 2003 ▶). This vector encodes a polypeptide with a hexahistidine tag at the N-terminus which facilitates purification. The pQE30-rspB construct was transformed into expression host Escherichia coli strain BL21 (DE3) and expression was performed in Luria–Bertani (LB) medium supplemented with 100 µg ml−1 ampicillin at 310 K until an optimum optical density (OD600 of 0.6) was reached. Following this, protein expression was induced with 1 mM IPTG (isopropyl β-d-1-thiogalactopyranoside) and incubation continued for an additional 4 h at 310 K. The cells were collected by centrifugation at 4000 rev min−1 for 20 min at 277 K. The cells were stored at 253 K until use.
2.2. Purification
For the preparation of a soluble protein fraction from a 1 l culture, the cell pellet was resuspended in 20 ml cold lysis buffer containing 20 mM Tris pH 8.0, 300 mM NaCl, 5% glycerol and 1 mM PMSF (phenylmethanesulfonylfluoride). It was then lysed by sonication on ice. The clear supernantant containing the soluble protein was collected by centrifugation at 10 000 rev min−1 at 277 K. The supernatant and pellet were checked on a 12.5% SDS–PAGE gel. The protein was found in the supernatant, which was used in subsequent purification steps. The first step of purification was performed by immobilized metal-affinity chromatography (IMAC), in which the supernatant was loaded onto an Ni–NTA column, washed with buffer (20 mM Tris pH 8.0, 300 mM NaCl and 5% glycerol) and eluted stepwise with a buffer containing increasing concentrations of imidazole. The elution fractions were checked on a 12.5% SDS–PAGE gel. The protein was eluted with 100 and 200 mM imidazole.
These two fractions were pooled, concentrated to approximately 2 ml and dialyzed overnight against 20 mM Tris pH 8.0, 300 mM NaCl and 5% glycerol to remove imidazole. The dialyzed sample was then loaded onto a Superdex-75 column (GE Healthcare Life Sciences). The peaks obtained were checked on a 12.5% SDS–PAGE gel. The fractions from the peak containing the protein were pooled and concentrated to around 500 µl using a Vivaspin microconcentrator (Sigma). The purity of the protein was estimated on a 12.5% SDS–PAGE gel and was found to be nearly homogenous. A final concentration of 55 mg ml−1 protein was obtained from the 1 l culture.
2.3. Preparation of selenomethionine derivative
RspB(31–348) contains ten methionine residues. A selenomethionine derivative was prepared by growing the same host cells in 1× M9 minimal medium (Sambrook et al., 1989 ▶) supplemented with the corresponding antibiotics. Before induction, the amino acids repressing methionine biosynthesis, l-Ile, l-Leu, l-Lys, l-Phe, l-Thr and l-Val, were added together with l-SeMet. Cell harvesting and purification took place as for the native protein.
2.4. Crystallization
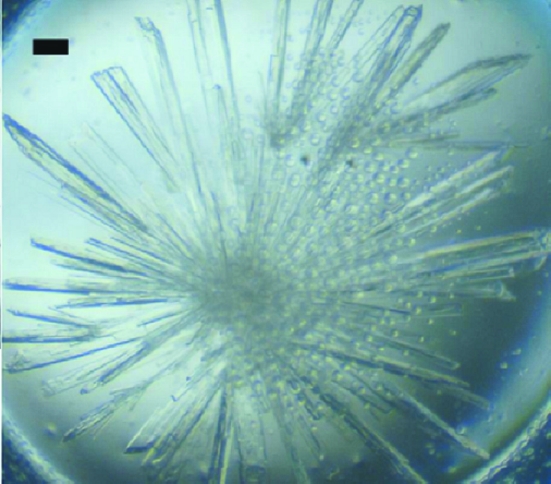
Crystallization experiments were carried out using the hanging-drop vapour-diffusion method with Hampton Research screens (Crystal Screen, Crystal Screen 2, PEG/Ion, PEG/Ion 2 and SaltRx) and home-made screens such as ammonium sulfate versus pH and polyethylene glycol versus pH. The condition that gave crystals was optimized to obtain diffraction-quality crystals. Initially, thin plate-like crystals were obtained which diffracted very poorly. Microseeds were prepared from these crystals and crystallization was carried out using these. Finally, drops containing 2 µl protein solution and 2 µl reservoir solution (35% PEG 2000 monomethyl ether, 100 mM sodium cacodylate pH 6.5 and 20 mM MgCl2) seeded with microseeds gave crystals suitable for data collection. The selenomethionine-incorporated rRspB(31–348) also gave crystals under the same condition as the native protein (Fig. 2 ▶).
Figure 2.
Long plate-like crystals of rRspB(31–348) from E. rhusiopathiae. The scale bar represents 0.1 mm.
3. Preliminary X-ray diffraction analysis and discussion
Several data sets were collected from a native rRspB crystal using our in-house X-ray diffraction facility: a Bruker Microstar copper rotating-anode generator operating at 60 mA and 45 kV with a MAR 345 image-plate detector. Generally, rRspB crystals diffracted to around 2.5 Å resolution; however, a complete data set could not be collected from a single crystal owing to the anisotropic nature of the diffraction, which was likely to be a consequence of the thin plate-like crystals. Merging different data sets gave a completeness of 98%, but the R value was high. The best native data set was obtained after merging data from two different crystals (details are given in Table 1 ▶). MAD data and a native data set were collected from selenomethionine-incorporated crystals on beamline XRD1, Elettra, Trieste, Italy. The incorporation of selenomethionine into the crystal was detected using X-ray fluorescence spectroscopy at the beamline. For data collection from both the native and selenomethionine-derivative crystals, the crystals were picked up in a loop and flash-cooled in a nitrogen-gas stream at 100 K. The diffraction images were indexed, integrated, merged and scaled using the AUTOMAR software package (Bartels & Klein, 2003 ▶).
Table 1. Data-collection statistics.
Values in parentheses are for the last resolution shell.
| Native | Selenomethionine derivative | |
|---|---|---|
| Unit-cell parameters (Å, °) | a = 46.19, b = 67.58, c = 102.31, β = 92.91 | a = 46.19, b = 66.65, c = 101.72, β = 94.11 |
| Space group | P21 | P21 |
| Resolution (Å) | 30–2.5 (2.59–2.50) | 30–2.2 (2.28–2.20) |
| Wavelength (Å) | 1.5418 | 1.000 |
| No. of crystals | 2 | 1 |
| Total No. of reflections | 51586 | 137821 |
| Unique reflections | 21673 | 39075 |
| Multiplicity | 2.53 (2.46) | 3.53 (3.69) |
| Completeness | 98.6 (98.8) | 99.3 (99.9) |
| Rmerge (%) | 12.21 (23.77) | 9.42 (30.70) |
| 〈I/σ(I)〉 | 3.0 (1.2) | 4.3 (1.2) |
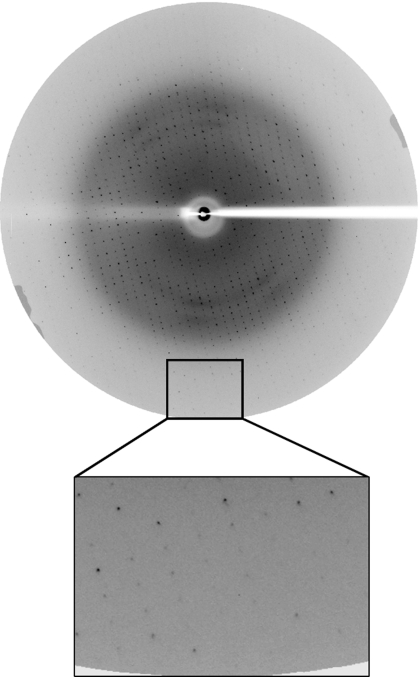
The selenomethionine-derivative crystal was used to collect a three-wavelength MAD data set of 360 frames each using a 1° oscillation step and an exposure of 20 s per frame. The crystal diffracted to 2.5 Å resolution, but MAD phasing was not successful owing to decay of the crystal during the course of data collection and poor anomalous signal. Native data were collected to 2.2 Å resolution from the selenomethionine-derivative rRspB crystal at a wavelength of 1 Å (Fig. 3 ▶). A total of 180 frames were collected using a 1° oscillation step and exposure of 20 s per frame; the data-collection statistics are summarized in Table 1 ▶. Both the native and selenomethionine rRspB proteins crystallized in the monoclinic space group P21. Assuming a calculated molecular weight of 37 255 Da for the protein and the presence of two molecules in the asymmetric unit, the resultant Matthews coefficient (Matthews, 1968 ▶) is 2.1 Å3 Da−1, corresponding to a solvent content of 41%. Attempts to obtain a molecular-replacement solution using the collagen adhesins Cna and Ace (PDB codes 2f68 and 2z1p, respectively) as the model did not yield any successful solution. Therefore, further MAD data sets will be collected in order to solve the structure of rRspB.
Figure 3.
Diffraction pattern of a selenomethionine-derivative crystal of rRspB(31–348). Diffraction spots are observed to a resolution of 2.2 Å.
Acknowledgments
KP thanks the Department of Science and Technology (DST), Government of India for financial support. KP thanks DST, India and the Ministry of Foreign Affairs/Sincrotrone Trieste, Italy for financial support to visit the Elettra synchrotron facility for data collection.
References
- Bartels, K. S. & Klein, C. (2003). The AUTOMAR Manual, v.1.4. Norderstedt, Germany: MAR Research GmbH.
- Gorby, G. L. & Peacock, J. E. Jr (1998). Rev. Infect. Dis.10, 317–325. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Navarre, W. W. & Schneewind, O. (1999). Microbiol. Mol. Biol. Rev.63, 174–229. [DOI] [PMC free article] [PubMed]
- Nerad, J. L. & Snydman, D. R. (1998). Infectious Diseases, edited by S. L. Gorbach, J. G. Bartlett & N. R. Blackhow, pp. 1755–1788. Philadelphia: WB Saunders Co.
- Norman, B. & Kihlstrom, E. (1985). Scand. J. Infect. Dis.17, 123–124. [DOI] [PubMed]
- Rich, R. L., Demeler, B., Ashby, K., Deivanayagam, C. C., Petrich, J. W., Patti, J. M., Narayana, S. V. & Hook, M. (1998). Biochemistry, 37, 15423–15433. [DOI] [PubMed]
- Rich, R. L., Kreikemeyer, B., Owens, R. T., LaBrenz, S., Narayana, S. V. L., Weinstock, G. M., Murray, B. E. & Hook, M. (1999). J. Biol. Chem.274, 26939–26945. [DOI] [PubMed]
- Robson, J. M., McDougall, R., van der Valk, S., Waite, S. D. & Sullivan, J. J. (1998). Pathology, 30, 391–394. [DOI] [PubMed]
- Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed., Book 3, p. A.3. New York: Cold Spring Harbor Laboratory Press.
- Scott, J. R. & Barnett, T. C. (2006). Annu. Rev. Microbiol.60, 397–423. [DOI] [PubMed]
- Shimoji, Y., Ogawa, Y., Osaki, M., Kabeya, H., Maruyama, S., Mikami, T. & Sekizaki, T. (2003). J. Bacteriol.185, 2739–2748. [DOI] [PMC free article] [PubMed]
- Venditti, M., Gelfusa, V., Castelli, F., Brandimarte, C. & Serra, P. (1990). Eur. J. Clin. Microbiol. Infect. Dis.1, 50–52. [DOI] [PubMed]
- Wood, R. L. (1999). Diseases of Swine, edited by A. D. Leman, B. E. Straw & W. L. Mengeline, pp. 419–430. Ames: Iowa State University Press.