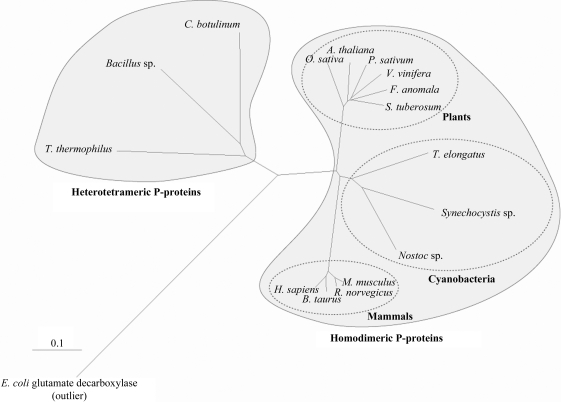
Figure 1.
Rooted tree of glycine decarboxylase proteins constructed from a ClustalX (Larkin et al., 2007 ▶) alignment of homodimeric and heterotetrameric glycine decarboxylase enzymes. The phylogenetic tree was constructed with the program TreeView (Page, 1996 ▶). To enable the alignment of enzymes with different oligomeric compositions, subunit 2 of the heterotetrameric P-proteins was fused to the carboxy-terminus of subunit 1. The tree shows that the Synechocystis P-protein clusters more closely with eukaryotic P-proteins than any of the heterotetrameric enzymes, including the T. thermophilus enzyme. E. coli glutamate decarboxylase (NP_287662) was defined as an outlier. P-protein sequences from the following eukaryotic species and bacterial isolates were used: Synechocystis sp. PCC 6803 (P74416), Thermosynechococcus elongatus Bp-1 (NP_682393), Nostoc sp. PCC 7120 (NP_488647), Oryza sativa (AAQ24377), Arabidopsis thaliana (BAE98954), Pisum sativum (P26969), Solanum tuberosum (O49954), Flaveria anomala (O49850), Vitis vinifera (XP_002279590), Mus musculus (NP_613061), Bos taurus (XP_869239), Homo sapiens (NP_000161), Rattus norvegicus (NP_001101053), Thermus thermophilus HB-8 (subunit 1, YP_143791; subunit 2, YP_143792), Bacillus sp. B14905 (subunit 1, ZP_01725792; subunit 2, ZP_01725793), Clostridium botulinum A strain ATCC 3502 (subunit 1, YP_001253239; subunit 2, YP_001253240). The scale bar shows the distance equal to 0.1 amino-acid substitutions per sequence position.