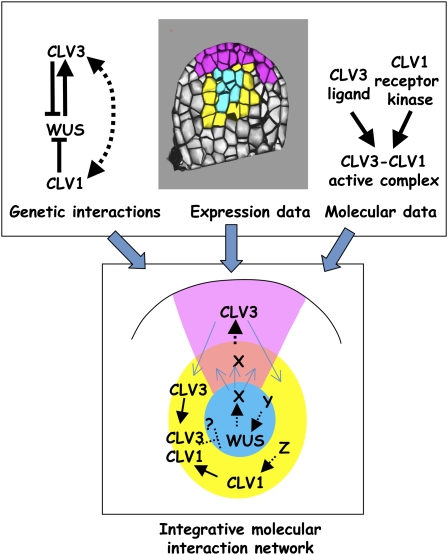
Figure 1.
Approach used for the reconstruction of an integrative molecular interaction network. We take as an example, the well-known WUS-CLV regulatory loop. Genetic interactions show that WUS is necessary for CLV3 activation and that it is itself repressed by CLV3 and CLV1, two genes acting in the same genetic pathway (for details see text). Expression data show that each of these three genes has a distinct and specific expression pattern within the meristem. From a molecular point of view, CLV3 encodes a ligand that is secreted and physically binds to the CLV1 receptor kinase. The protein complex formed by this interaction is necessary to restrict the expression of WUS in the organizing center. The structure of the molecular regulatory network is given by the molecular interactions between the entities of this network. For each element differentially localized among regions, the network must reproduce the active or inactive state of the corresponding element and the incident interactions must therefore be able to account for these different states. The network can then be used to run simulations and see which stable states can be obtained. In parallel, expression data for the molecular elements of the network can be compiled and superposed on a virtual meristem to identify zones of cells that coexpress the same set of elements. In principle, the stable states of the network should correspond to the coexpression zones observed. Lastly, genetic interaction data, which give information on the dynamic of the network but not on the molecular trajectories, should be explained by the molecular network. Due to the lack of available molecular interaction data, the problem is in fact to build the network. In the case of WUS-CLV, we not only lack molecular interactions (for example, is the repression of WUS by CLV3-CLV1 direct?) but we also cannot explain WUS and CLV1 expression pattern since their activators are unidentified yet. In addition, it is likely that there exists an intermediate messenger for the activation of CLV3 by WUS since their expression pattern does not overlap. We can therefore introduce hypothetical molecular interactions (dotted arrows) and see if it is consistent with both expression and genetic data. These hypothetical interactions can then be tested experimentally and used either to confirm the validity of the network, or to introduce alternative interactions. This is an iterative process that requires the use of computer modeling to run simulations and to generate predictions.