Humpty Dumpty, a traditional English nursery rhyme:
Humpty Dumpty sat on a wall,
Humpty Dumpty had a great fall.
All the king's horses,
And all the king's men,
Couldn't put Humpty together again.
One definition of systems biology is the modeling of the relationships, or networks, within and/or between large-scale data sets. As scientists' knowledge of living systems has grown, it has become increasing difficult to understand relationships between all the components of a biological network by intuitive approaches alone. The value of systems biology is that it (1) helps us to organize information about complex systems; (2) reveals hidden content, or “emergent properties,” of the system that would not be deduced from a study of the separate parts of the system; and (3) provides predictions of how the system would behave under conditions that have not yet been observed. Such predictions can then be used to inform and direct wet bench experiments.
THE SYSTEMS OF SYSTEMS BIOLOGY
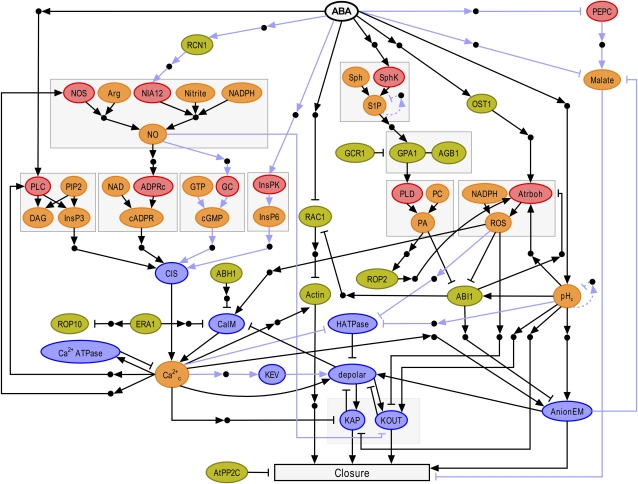
There are many different types of biological networks that can be investigated using systems biology approaches. Some systems are typified by a certain type of biological molecule (e.g. all of the transcription factors that are activated by cold stress or all of the proteins that are phosphorylated by a particular kinase). Other systems are studied at a particular level of organization, such as the subcellular, cellular, tissue, organ, whole plant, or ecosystem. Systems biology can yield insights into both developmental processes, such as the classic ABC model of flower development (Weigel and Meyerowitz, 1994), and physiological processes, such as pathogen signaling networks (Agrawal et al., 2004). This Update focuses on the systems biology of rapid cellular signaling, using as an example abscisic acid (ABA) signaling in guard cells (Figs. 1 and 2). Accordingly, the goal of this Update is not to review ABA regulation of stomatal movements per se but rather to use this phenomenon to illustrate some general aspects of systems biology approaches toward modeling cellular signaling cascades. Figure 2 shows a signaling network for ABA-induced stomatal closure (Li et al., 2006) that can be used as a reference point for the concepts discussed in this Update. Our knowledge of ABA signaling has progressed substantially since this figure was first published in 2006, and some of the major advances are also highlighted.
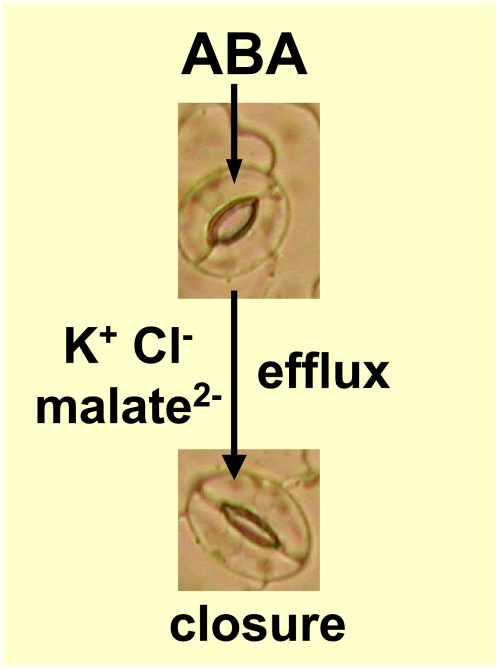
Figure 1.
ABA-stimulated solute efflux drives stomatal closure.
Figure 2.
Network of ABA-induced stomatal closure. (Figure and figure legend are reproduced from Li et al. [2006].) The color of each node represents its function: enzymes are shown in red, signal transduction proteins are green, membrane transport-related nodes are blue, and secondary messengers and small molecules are shown in orange. The full names of network components corresponding to each abbreviated node label are indicated below. Small black circles represent putative intermediary nodes mediating indirect regulatory interactions. Arrowheads represent activation, and short perpendicular bars indicate inhibition. Light blue lines denote interactions derived from species other than Arabidopsis, and dashed light blue lines denote inferred negative feedback loops on pH and S1P. Nodes involved in the same metabolic pathway or protein complex are bordered by gray boxes; only those arrows that point into or out of the boxes signify information flow (signal transduction). ABI1/2, Protein phosphatase 2C ABI1/2; ABH1, mRNA cap-binding protein; Actin, actin cytoskeleton reorganization; ADPRc, ADP-Rib cyclase; AGB1, heterotrimeric G protein β-subunit; AnionEM, anion efflux at the plasma membrane; AtPP2C, protein phosphatase 2C; Atrboh, NADPH oxidase; CaIM, Ca2+ influx across the plasma membrane; Ca2+ ATPase, Ca2+ ATPases and Ca2+/H+ antiporters responsible for Ca2+ efflux from the cytosol; Ca2+c, cytosolic Ca2+ increase; cADPR, cyclic ADP-Rib; cGMP, cyclic GMP; CIS, Ca2+ influx to the cytosol from intracellular stores; DAG, diacylglycerol; ERA1, farnesyl transferase ERA1; GC, guanyl cyclase; GCR1, putative GPCR; GPA1, heterotrimeric G protein α-subunit; HATPase, H+-ATPase at the plasma membrane; InsPK, inositol polyphosphate kinase; InsP3, inositol-1,4,5-trisphosphate; InsP6, inositol hexakisphosphate; KAP, K+ efflux through rapidly activating K+ channels (AP channels) at the plasma membrane; KEV, K+ efflux from the vacuole to the cytosol; KOUT, K+ efflux through slowly activating outwardly rectifying K+ channels at the plasma membrane; NOS, nitric oxide synthase; NIA12, nitrate reductase; NO, nitric oxide; OST1, protein kinase OST1; PA, phosphatidic acid; PC, phosphatidylcholine; PEPC, phosphoenolpyruvate carboxylase; PIP2, phosphatidylinositol 4,5-bisphosphate; PLC, phospholipase C; PLD, phospholipase D; RAC1, small GTPase RAC1; RCN1, protein phosphatase 2A; ROP2, small GTPase ROP2; ROP10, small GTPase ROP10; SphK, sphingosine kinase; Sph, sphingosine; S1P, sphingosine-1-phosphate.
STOMATAL GUARD CELLS
Guard cells are morphologically distinct cells that are located in pairs in the epidermes of aerial plant parts, where they define the apertures of stomatal pores, through which land plants take up carbon dioxide for photosynthesis but also, inevitably, lose water vapor (Fig. 1). Guard cells have evolved exquisite mechanisms to sense and respond to endogenous and environmental stimuli such as hormones, light, carbon dioxide concentrations, temperature, humidity, and plant water status, with the result that stomatal apertures are narrowed under water-limiting conditions and widened when conditions for photosynthesis are optimal. Appropriate guard cell function is vitally important for the survival of land plants and thus has direct implications for the production of adequate food and fuel to provision the burgeoning human population. Moreover, considering the list of stimuli to which guard cells respond, it can be anticipated that guard cell responses will both be affected by and will feed back to affect global environmental parameters such as planetary temperature and atmospheric concentrations of greenhouse gases such as water vapor and CO2 (Kirschbaum, 2004). Indeed, it has been predicted that stomatal closure resulting from a doubling of atmospheric CO2 concentrations would decrease water vapor loss through stomata to such an extent that continental runoff of freshwater would increase significantly (Betts et al., 2007).
In addition to the relevance of guard cells to the survival of terrestrial organisms, guard cells have also become a premier model system for the study of signaling at the cellular level, in part due to their accessibility and the availability of techniques to dynamically monitor their cellular output. Because guard cells are located in the outermost cell layer of the plant, the epidermis, the effects of a stimulus can be readily and directly monitored in real time at several different levels. First, at the subcellular level, electrophysiological techniques allow the monitoring of stimulus-induced changes in ion fluxes across the guard cell membrane. Changes in the concentrations of ions and other solutes result in osmotically driven water fluxes across the guard cell membrane that in turn cause cell swelling or shrinking, resulting in stomatal opening or stomatal closure, respectively (Fig. 1). Imaging techniques also allow monitoring of changes in the concentrations of signaling ions, particularly H+ and Ca2+, in real time. Second, responses of individual guard cells to a stimulus can be quantified by direct observation of stomatal apertures with a microscope. Third, effects of a stimulus on stomatal apertures can be monitored at the integrated whole leaf level by measurements of the rate of water vapor loss (transpiration) from the leaf. From assays such as these, we know that when plants are drought stressed and guard cells are exposed to elevated ABA levels, ABA stimulates the efflux of K+, Cl−, and malate2− from the guard cells as well as Suc efflux or metabolism under some conditions. The resultant decrease in intracellular concentrations of these major solutes drives water efflux, thus shrinking guard cell volume and narrowing stomatal apertures (Fig. 1). In addition to the above real-time measurements, the ability to isolate large populations of highly pure guard cells has enabled the determination of guard cell transcriptomes and guard cell proteomes (Leonhardt et al., 2004; Yang et al., 2008; Zhao et al., 2008; Zhu et al., 2009).
COLLECTING INFORMATION: IDENTIFYING NODES
To model a cellular signaling system such as ABA-induced stomatal closure, the first steps are to identify the components of the system and to define the relationships between them. In a systems biology analysis, the components of the system are depicted as nodes and their relationships with each other are depicted as edges (Fig. 2), thus resulting in the assembly of a network. Edges can be either directional, indicating a specific direction of information flow (e.g. as in a signal transduction network), or nondirectional (e.g. as in the interaction of two scaffolding proteins). Edges can either be positive, indicating activation, or negative, indicating inhibition.
Identification of the nodes of cellular signaling systems has benefited from the powerful tools of forward and reverse genetics. For example, forward genetic screens in Arabidopsis (Arabidopsis thaliana) for ABA effects on seed germination and seedling growth led to the identification of ABA-insensitive mutants such as abi1-1 (Koornneef et al., 1984). When the abi1-1 mutant was subsequently analyzed for guard cell properties, it was found that this dominant mutation conferred ABA hyposensitivity to guard cell responses as well (Pei et al., 1997; Allen et al., 1999). Cloning of ABI1 showed that this gene encodes a PP2C-type protein phosphatase (Meyer et al., 1994; Leung et al., 1997), indicating that protein de/phosphorylation plays an important role in ABA signaling. Subsequent identification of recessive abi1 alleles showed that the ABI1 gene product actually functions as a negative regulator of ABA signaling (Merlot et al., 2001).
Gene identification based on forward genetic screens such as these has resulted in the identification of numerous guard cell ABA signaling components. It is interesting, however, that the majority of these mutants were initially identified by screening for a non-guard cell aspect of ABA physiology, in particular ABA inhibition of seed germination (Wang et al., 2004), and then the mutant was subsequently assessed for a possible guard cell phenotype. This is most likely due to the fact that stomatal apertures are microscopic, change rapidly, and are also affected by a plethora of stimuli, making it a challenge to devise a robust forward genetic screen of guard cell physiology. In recent years, however, infrared thermography has been used to screen for mutants with altered leaf temperature resulting from defects in transpirational cooling, and this approach has resulted in the identification of genes that impact guard cell responses to ABA, humidity, and CO2 (Merlot et al., 2002, 2007; Wang et al., 2004; Xie et al., 2006; Negi et al., 2008).
Reverse genetic approaches are also potent tools in the identification of cellular signaling components. In reverse genetics, a gene is first identified to be of interest, and then the impact of the knockout of this gene on the relevant system is assessed in order to decide whether or not the gene product should be included as a node in that network. A gene may be deemed to be of interest because it has homology to a protein of relevant function in another organism. For example, certain phosphorylated lipid metabolites have been implicated as secondary messengers in guard cell ABA signaling, and candidates for the genes that encode the associated kinases were identified in the Arabidopsis genome based on homology to such kinases in mammals (Worrall et al., 2008). A gene may also be deemed to be of interest because it has been shown to function in an analogous signaling pathway in a different cell type in the same organism. In Arabidopsis, for example, phototropin genes were first identified to encode photoreceptors for blue light-induced phototropic bending (Christie et al., 1998); subsequently, double knockout mutants of phot1 and phot2 were shown to be impaired in blue light-induced stomatal opening, thus implicating PHOT1 and PHOT2 as guard cell blue light photoreceptors (Kinoshita et al., 2001).
A gene may also be deemed to be of interest because its transcript or protein product is highly abundant or highly regulated in the cellular system of interest. For example, the Arabidopsis GORK gene is now known to encode a K+ channel protein that mediates K+ efflux during stomatal closure. gork knockouts were initially targeted for investigation because of high GORK expression levels in wild-type guard cells, coupled with the observation that the GORK protein exhibits K+ channel activity when expressed in heterologous systems (Hosy et al., 2003). At the protein level, a recent initial identification of the Arabidopsis guard cell proteome (Zhao et al., 2008) showed that the myrosinase enzyme TGG1, which produces isothiocyanates by catalyzing the hydrolysis of glucosinolates, is one of the most abundant guard cell proteins. Reverse genetic analysis then revealed that TGG1 participates in guard cell responses to ABA (Zhao et al., 2008; Islam et al., 2009), thus adding another function to this protein's repertoire, in addition to its known roles in herbivore response. Although there are still only a few reports on the guard cell transcriptome and proteome (Leonhardt et al., 2004; Yang et al., 2008; Zhao et al., 2008; Zhu et al., 2009), these publications nevertheless provide fertile ground for the mining of additional candidate nodes to be assessed with respect to stomatal function.
Once a gene (product) has been confirmed as a node in a cellular signaling system, coexpression analysis of other transcriptomic (or proteomic) data sets can be used to identify additional candidates that coexpress with the first gene/protein both in guard cells and in other tissues or conditions and, thus, are themselves implicated as having a functional relationship with the first gene/protein (Albert and Albert, 2004; Aoki et al., 2007). Such approaches have not yet been applied to the guard cell system, but they could be particularly useful in prioritizing the wet bench assessment of genes of unknown function.
Signaling networks employ not only proteins but also ions and metabolites as secondary messengers. Fluorescent reporters of ion concentration have implicated Ca2+ and H+ ions as important secondary messengers in single cell systems such as guard cells and pollen tubes. In particular, fluorescent reporters of cytosolic Ca2+ concentration have illuminated the role of Ca2+ as a secondary messenger in guard cell ABA responses, and elevation of cytosolic pH is also an important component in guard cell ABA signaling. Only a few single-cell plant systems, such as trichomes (Schilmiller et al., 2008), have been subjected to detailed metabolite analysis to date. At present, pharmacological assays predominate as a tool to identify metabolite involvement in cellular signaling pathways, both for guard cells and for other cell types. In guard cells, lipid-based signals, cyclic nucleotides, cADPR, reactive oxygen species (ROS), and nitric oxide all have been implicated in ABA signaling by pharmacological approaches. In some cases, this pharmacological evidence is supported by strong biochemical or molecular genetic analyses, while in other cases, such evidence is still incomplete. Some of these issues will be resolved once large-scale metabolomics approaches are applied to the analysis of guard cell signaling cascades.
After assembling information on candidate nodes, the researcher must decide what standard of proof will be required for inclusion of a node in a network. Will pharmacological evidence be considered as sufficient, or is genetic and/or biochemical information required? Is pharmacological evidence less convincing because of the known lack of specificity of pharmacological interventions? Or is pharmacological evidence actually more relevant for the study of rapid physiological responses such as stomatal closure because it is a short-term manipulation and thus less likely to have pleiotropic or developmental effects that could contribute to altering the overall cellular status or “set point” of the cell? Once a standard of proof is decided on, it obviously should be applied uniformly to all the nodes under consideration for inclusion in the network.
COLLECTING INFORMATION: DRAWING EDGES
Once the set of nodes has been identified, the relationships between them must be formalized in order to actually draw the network. Again, there are several approaches that can yield useful information. Geneticists have long employed epistasis analysis to determine the relationship between genes/gene products, and such analyses provide one mechanism to place two proteins within the same branch of a signaling network or to determine that the proteins are functioning independently, additively, or synergistically. Epistasis analysis, however, does not determine whether proteins situated in the same path of a network are actually adjacent to each other in the pathway. In the model of Figure 2, whenever indirect evidence positioned two nodes as adjacent in a pathway but direct evidence that the two nodes physically interacted was lacking, a small black circle (representing the possibility of additional, intermediate nodes) was added.
Evidence for direct node-node interactions can be provided by various protein-protein interaction assays or by detailed biochemical analyses that determine the interrelationships between two proteins or between a protein and a cofactor or metabolite. For example, it had been observed that mutation of the ABA-activated protein kinase, OST1, impaired ABA-stimulated production of ROS in guard cells and resulted in other phenotypes that were similar to those observed when the NADPH oxidases AtrbohD and AtrbohF were knocked out (Mustilli et al., 2002). Based on such observations, Li et al. (2006) placed OST1 upstream of Atrboh in the network of Figure 2. Recently, mass spectrometric analysis of Atrboh phosphorylation status and in vitro kinase assays have shown that AtrbohF is indeed a phosphorylation target of OST1 (Sirichandra et al., 2009), thus supporting the positioning of OST1 and Atrboh in Figure 2.
When drawing edges between two nodes, the researcher must again decide what standard of proof to apply. For example, there are a variety of methods for determining protein-protein interaction, some of which are performed in vitro, others in yeast, and others in planta (Lalonde et al., 2008). Will demonstration that two proteins interact in vitro be considered sufficient, will the two proteins need to be shown to interact by two or more independent approaches, or will the most rigorous (and technically difficult) standards of proof, such as demonstration of in planta interaction via Förster resonance energy transfer, be required?
The most informative signaling networks are those with edges that indicate the direction and type of information flow; so in building a signaling network, the researcher needs to identify not only which nodes are upstream and which are downstream but also which are activating and which are inhibitory. Again, genetic and biochemical approaches yield useful information, with the caveat that directionality is not indicated by all assay methods. For example, yeast two-hybrid or coimmunoprecipitation analyses show that two proteins interact but are not sufficient to indicate which protein is upstream and which is downstream in a signaling cascade, nor do such data indicate whether the interaction is positive or negative.
There are some simplifying assumptions that can be applied to aid the process of drawing edges. One overall simplifying assumption is that of parsimony (i.e. that one should construct the simplest possible network that is in accordance with the experimental data). For example, if A is known to promote both B and C and C is known to promote the A → B process, then one can infer the relationship A → C → B. Other inferences that can be incorporated to satisfy parsimony are given by Li et al. (2006) and Albert et al. (2007). A software package, NET-SYNTHESIS (http://www.cs.uic.edu/∼dasgupta/network-synthesis/), is available that takes the causal relationships among components as defined by the user and incorporates such inferences to draw the most parsimonious network (Kachalo et al., 2008).
ANALYZING NETWORK PROPERTIES
Once the structure of a network has been finalized, its properties can be assessed. Emergent properties are those that could not be deduced by studying individual components in isolation (Bhalla and Iyengar, 1999). In Figure 2, the ultimate emergent property is stomatal closure, but there are also important properties within a network that can be considered as emergent. Some general aspects of networks that can be considered as emergent properties are motifs, modules, and scale. Negative and positive feedback loops are familiar examples of motifs. For example, in the network of Figure 2, cytosolic Ca2+ elevation, which activates the Ca2+ extruding ATPase, which in turn decreases cytosolic Ca2+ levels, is an example of a negative feedback loop (Ca2+c → Ca2+ ATPase ⊣ Ca2+c), while cytosolic Ca2+ activation of the enzyme phospholipase C (PLC), leading to production of inositol 1,4,5 trisphosphate (InsP3), which stimulates Ca2+ release from intracellular stores, which elevates cytosolic Ca2+ (Ca2+c → PLC → InsP3 → Ca2+c), is an example of a positive feedback loop. Motifs may or may not occur in modules, which are subnetworks within the main network that have high interconnectivity (numerous edges) within the module and low interconnectivity (few edges) between the module and the rest of the network.
In many types of networks, including biological networks, it has been observed that there is typically a large range in the number of edges that the nodes of the network exhibit; thus, the network cannot be described adequately by a “typical” or average number of connections. Such networks are said to be scale free (Albert et al., 2000; Jeong et al., 2000). The network of Figure 2 is not large enough to either verify or disprove its scale-free nature. In scale-free networks, there will be a minority of nodes that have a much higher than average number of connections. Such highly connected nodes are called hubs. Hubs of cellular networks can be composed of any type of biological molecule, including proteins, metabolites, nucleic acids, and so on, depending on the specific network under consideration. It is easy to envision that knockout of hubs is, on average, more likely to have a deleterious effect on a system than knockout of nodes with only a few connections. This intuition has been confirmed by studies of gene lethality in yeast (Jeong et al., 2001; He and Zhang, 2006).
MODELING: CHOOSING A MODELING FRAMEWORK
One major goal of systems biology is to develop models that have predictive value. Once a network has been assembled, the type of modeling that can be performed with it is dependent on the depth of knowledge that is available about the system and, particularly, on whether that knowledge is quantitative or qualitative. If detailed quantitative and kinetic information is available on the status of the nodes, then a continuous model can be developed using ordinary differential equations or partial differential equations. A good introduction to these types of models is provided by Eungdamrong and Iyengar (2004). For guard cell biology, while available techniques readily allow the collection of quantitative information at the level of the output node (e.g. the size of the stomatal aperture or the rate of transpirational water loss over time), there are only a handful of cases in which quantitative information is available regarding the status of the internal nodes in this network. Quantitative information is available from electrophysiological measurements of ion channel activity, and from fluorescent imaging of Ca2+ elevations and oscillations in response to ABA treatment. There is some quantitative information on other aspects, such as the rates of production of ROS and some lipid metabolites, but the information is insufficient to allow a completely continuous modeling approach.
Fortunately, there are other methods of network analysis that can be applied to make predictions in the absence of detailed quantitative information. Hybrid models can be applied in which some variables are continuous and others discrete, or a fully discrete model can be utilized. In a discrete model, nodes are restricted to particular states (e.g. fully on, partly on, or off). The most reduced form of a discrete model is a Boolean model, in which nodes are characterized as either on or off depending on the status of the upstream nodes and their relationship to those nodes (Albert and Wang, 2009). To date, ABA-induced stomatal closure has only been modeled with the Boolean approach (Li et al., 2006). Since the status of our knowledge of guard cell signaling is fairly typical of the status of our knowledge of other cellular signaling phenomena both in plant and nonplant systems (in fact, we have more complete knowledge of guard cell signaling than we do for many other single cell signaling cascades), Boolean modeling is discussed in more detail below. A primer on this method is also available (Assmann and Albert, 2009).
To perform Boolean modeling, logic rules that define the on (active) state of each node are first defined, using the Boolean operators “and,” “or,” and “not.” For example, when ABA activates the protein kinase OST1, the model of Figure 2 predicts that this will in turn activate the NADPH oxidases (Atrboh): since OST1 is the only input into Atrboh, Atrboh will be active (on) whenever OST1 is active. For nodes with more than one input, the rules can become more complicated. For example, as shown in Figure 2, there are four secondary messengers (InsP3, cADPR, cGMP, and InsP6) implicated in stimulating Ca2+ release from internal stores (node CIS [see Fig. 2 legend]). What is the most biologically accurate way to define the active state of node CIS? Possibilities include (InsP3 or cADPR or cGMP or InsP6), (InsP3 and cADPR and cGMP and InsP6), and various mixed combinations. The experimenter's goal is to define each rule so as to incorporate existing knowledge as well as possible. In some instances, there will be conflicting or insufficient data in the literature. Thus, building these rules requires not only logic but extensive application of the scientist's expertise.
Once Boolean rules have been designated for each node, there are two more decisions to make before modeling can begin. The first decision concerns how to set the initial status of the internal nodes. When modeling an irreversible developmental process, it may be a reasonable simplification to assume that all nodes downstream of the input node are inactive (off) until the input is turned on. However, for a reversible physiological process, such as stomatal closing and opening, this assumption seems likely to be invalid, absent evidence to the contrary, and so alternative approaches are desirable. One alternative is to randomly set the initial status of the internal nodes. Thus, when ABA-induced stomatal closure was simulated using the network of Figure 2, Li and colleagues (2006) modeled 10,000 in silico stomata, with the on or off status of each internal node randomly chosen anew for each in silico stoma, in order to obtain a prediction of network behavior (although 10,000 does not cover all random combinations of the node state space, results did not differ when 100,000 stomata were modeled, indicating that a sample size of 10,000 was sufficient). Another alternative is to incorporate prior knowledge to set the initial status of the internal nodes for which such information is available, while allowing the initial status of the rest of the internal nodes to be defined randomly. For example, in the network of Figure 2, if it were known that phosphoenolpyruvate carboxylase (PEPC) was always active in malate production in the absence of ABA, then the PEPC node could always initially be set to on for the modeling process.
Once an initial status of all the internal nodes has been established, randomly or otherwise, the input node can be turned on and the propagation of the signal through the network can be assessed. The second decision involves choosing how such propagation will be modeled (Albert and Wang, 2009). One possibility is to take recurring “snapshots” of the status of the network; in each snapshot, every node is updated simultaneously based on the information flowing into it from its inputs. For example, for Figure 2, the snapshot at t = 0 would show the ABA node as off and each internal node randomly set either to on or off. The second snapshot at t = 1, “instantaneously” following ABA application, would have the ABA node on. The next snapshot would show what had happened as a consequence of the ABA node turning on: for example, if all internal nodes were initially off, then in this next snapshot, nodes RCN1, InsPK, SphK, OST1, and pHc (for definitions, see Fig. 2 legend), would all get turned on, since all these nodes are immediately downstream of the ABA node, have as their input only the ABA node, and are positively regulated by ABA. The nodes malate and PEPC are also immediately downstream of ABA and exclusively regulated by ABA, but this regulation is inhibitory, so these nodes would turn off, if on, or remain off if they were off in the initial condition. The next snapshot in the continued presence of ABA would show the consequences of the new status of the seven above-mentioned nodes, etc. This process is referred to as synchronous updating, because all nodes have their status changed simultaneously, based on the prevailing status of their upstream neighbors.
In the absence of loops, synchronous modeling results in information percolating from level to level in the network, with information flowing most quickly in the shortest paths (i.e. the paths with fewest nodes). For developmental processes, this might be reasonable as a first approximation. However, for reversible physiological processes, this approximation is less likely to reflect biological reality. An alternative possibility is to perform asynchronous updating, in which the internal nodes are updated sequentially in a randomly chosen order, until every internal node has been updated once (Albert and Wang, 2009). Whether or not the network is then in a state such that the output node (in Figure 2, the output node is “closure”) will turn on is then assessed. Dynamic modeling with asynchronous (random) update is how Li et al. (2006) modeled ABA-induced stomatal closure in the 10,000 in silico stomata using the network of Figure 2. However, if some knowledge concerning rates of signal propagation along certain paths of the network is available, it is also possible to impose restrictions on the asynchronous update process such that, for example, certain nodes always are updated before certain other nodes in the network, thus allowing a closer approximation of the true dynamics of the signaling system.
After a number of rounds of update, the output of a Boolean network will either reach a steady state or start to cycle. In the case of the ABA network of Figure 2, outputs reached steady state within eight rounds of updates. It can be readily understood that this type of modeling requires programming for all but the simplest networks. Fortunately, Albert and colleagues (2008) have developed online software tools to facilitate this process.
MODELING: EVALUATING THE NETWORK AND MAKING PREDICTIONS
The first goal in running simulations such as those described above is to assess whether the model reflects the reality of the wild-type response of the cellular signaling system. In the case of Figure 2, the model satisfied this condition in that it did indeed predict ABA-induced stomatal closure. It has also been observed that many biological signaling systems are robust; that is, the outcome is, to some extent, resilient against perturbation. The signaling network of Figure 2 was found to be robust against random rewiring, a typical approach to evaluate robustness (Shen-Orr et al., 2002). Had robustness not been observed, this would have suggested that reevaluation of the network structure and Boolean rules was warranted or that insufficient information was available to apply this modeling approach.
If the model satisfies initial requirements, then one can proceed to use the model to simulate interesting conditions and make predictions regarding which of the simulated changes are most likely to affect the output. For ABA-induced stomatal closure, 65% of single node knockouts were predicted not to alter ABA-induced stomatal closure, which is also consistent with a robust system; this percentage dropped to 38% for double node knockouts (Li et al., 2006). These simulations accordingly lead to specific hypotheses as to which nodes in the system represent particular vulnerabilities. Such predictions can then be tested experimentally using reverse genetic and other methods. Inevitably, not all predictions will be supported, and this new information can be incorporated into a revised model. Thus, experiments that are designed based on such simulations provide information to improve the model, which then leads to resimulation and more predictions, in an iterative positive feedback loop. A situation that both the modeler and the reader must live with is that, especially in these early days of cellular systems biology, models are likely to become obsolete almost as soon as they are published. As one article states, “…models such as these should not be considered as definitive descriptions of networks within the cell, but rather as one approach that allows us to understand the capabilities of complex systems and devise experiments to test these capabilities” (Bhalla and Iyengar, 1999). On the other hand, if the model is robust, much of the new information, while adding biologically important detail, may not substantively change the network properties or model outcome.
RECENT DISCOVERIES IN GUARD CELL ABA SIGNALING
Guard cell signaling is a rapidly advancing area of research, and it is not possible within the constraints of an Update article to cover more than a small fraction of the recent publications in the field. This section very briefly summarizes some of the new information on guard cell signaling, focusing only on information that is relevant to ABA-induced stomatal closure, and thus could be used in the future to generate a revised network for this process.
Channels and transporters for ions and other solutes are the effectors of the membrane potential and water potential changes that drive stomatal movements. In response to ABA, inhibition of H+-ATPase-based H+ extrusion and activation of Ca2+ influx channels and anion efflux channels all contribute to membrane depolarization, which activates the voltage-regulated K+ channels through which K+ efflux occurs; this solute efflux results in water efflux and stomatal closure (Fig. 1). Previously identified transport proteins of the guard cell tonoplast and plasma membrane have been described in detail in recent review articles (Pandey et al., 2007; Ward et al., 2009). Recent advances have correlated additional gene products with the electrophysiological and transport signatures of channel and pump activity. For example, one of a dozen Arabidopsis H+-ATPases, AHA1, has been particularly implicated as a specific target for down-regulation during ABA-induced stomatal closure (Merlot et al., 2007). Glutamate receptors and cyclic nucleotide-gated channels have been implicated as channel types that could mediate Ca2+ influx across the plasma membrane (Ali et al., 2007; Cho et al., 2009). There is now strong evidence that the gene SLOW ANION CHANNEL ASSOCIATED1 encodes a slow anion efflux channel of guard cells (Negi et al., 2008; Vahisalu et al., 2008; Geiger et al., 2009; Lee et al., 2009). Solute efflux across the guard cell membrane must be preceded by solute release from the vacuole, and the gene TPK1 has recently been shown to encode a channel that mediates K+ flux across the guard cell tonoplast (Bihler et al., 2005; Gobert et al., 2007).
One of the most important recent discoveries is the identification of membrane-associated and soluble ABA receptors. Membrane-associated receptors, GTGs, interact with and are regulated by the Gα protein GPA1 (Pandey et al., 2009), and a plasma membrane-localized Leu-rich repeat receptor-like kinase, RPK1, has also been implicated in ABA perception (Osakabe et al., 2005). The soluble PYR1/PYL/RCAR ABA receptors (Ma et al., 2009; Park et al., 2009; Santiago et al., 2009a, 2009b) interact with and inhibit the negative regulator ABI1 and related PP2C protein phosphatases (Saez et al., 2004, 2006; Kuhn et al., 2006), and ABA activates the protein kinase OST1 by virtue of inhibiting these PP2C phosphatases (Yoshida et al., 2006; Fujii et al., 2009; Santiago et al., 2009b; Umezawa et al., 2009). Thus, while in the model of Figure 2 ABI1 is both a target of inhibition by ROS (Meinhard and Grill, 2001) and upstream of ROS by virtue of the observation that the dominant abi1-1 mutation inhibits ABA-induced ROS production (Murata et al., 2001), it now seems plausible that the inhibitory effect of ABI1 on Atrboh should be positioned farther upstream in that branch of the signaling cascade (i.e. upstream of OST1). It will be interesting to ascertain whether members of these new receptor types interact with any of the other seven nodes that are positioned adjacent to ABA in Figure 2 by virtue of the fact that, at the time the network was drawn, there was no evidence for components acting between these nodes and ABA. Note that in Figure 2 all of these nodes have black circles immediately above them, indicating that there was also no evidence for direct interaction of ABA with any of the nodes.
Further evidence for the central importance of protein de/phosphorylation in guard cell physiology comes from the latest reverse genetic studies on several types of protein kinases. Members of mitogen-activated protein kinase cascades have been implicated in the regulation of guard cell physiology as well as guard cell development (Colcombet and Hirt, 2008). The CBL-interacting protein kinase, CIPK23, regulates K+ channels (Xu et al., 2006), and regulation of CIPK23 by the concerted action of the calcineurin B-like proteins CBL1 and CBL9 has been implicated in stomatal aperture control by ABA (Cheong et al., 2007). The CBL proteins interact with target CIPKs upon Ca2+ binding, and certain members of the large family of plant calcium-dependent protein kinases (CDPKs) also modulate guard cell ABA signaling in a Ca2+-dependent manner. The CDPKs CPK6 and CPK3 are required for full ABA activation of slow anion channels and Ca2+-permeable channels of guard cells (Mori et al., 2006). The CDPKs CPK4 and CPK11 phosphorylate transcription factors ABF1 and ABF4, and ABF4 is known to interact with ABA response elements to regulate gene expression (Kang et al., 2002; Zhu et al., 2007). Down-regulation of certain AP2-type (Song et al., 2005) and MYB transcriptional regulators (Cominelli et al., 2005; Jung et al., 2008; Ding et al., 2009) has also been shown to alter ABA responsiveness and drought tolerance by targeting the expression of stress signaling elements. While these studies contribute new nodes to the network of ABA-induced stomatal closure, other recent studies further address relationships among the extant nodes (i.e. the “wiring” of the network; Bright et al., 2006; Fan et al., 2008; Gonugunta et al., 2008; Perera et al., 2008; Saito et al., 2008; Worrall et al., 2008; Zhang et al., 2009).
CONCLUSION
These are still early days for cellular systems biology. For example, the network of Figure 2 was, at the time, the most complex network to have been modeled by Boolean modeling with asynchronous update. Yet, ensuing rapid progress in guard cell signaling has provided a number of new nodes and edges, with still more to come. Thus, we are quite some time away from having a comprehensive model of ABA-induced stomatal closure. In addition, since ABA is just one facet of the multisensory capacities of the guard cell, in order to fully predict stomatal behavior it will be necessary to integrate the ABA signaling network with those for other hormones and for light, CO2, humidity, temperature, and pathogen sensing. Nevertheless, the promise of systems biology is that the wealth of information being made available by reductionistic approaches can be synthesized to yield a greater understanding of biological systems. In other words, after decades of taking apart Humpty Dumpty to figure out his components, systems biology offers the kings' soldiers a path by which Humpty might indeed eventually be put together again.
Acknowledgments
I thank Prof. Réka Albert for helpful comments on the manuscript, and I apologize to those many authors whose work was not cited owing to space limitations.
This work was supported by the National Science Foundation and the U.S. Department of Agriculture.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Sarah M. Assmann (sma3@psu.edu).
References
- Agrawal V, Zhang C, Shapiro AD, Dhurjati PS (2004) A dynamic mathematical model to clarify signaling circuitry underlying programmed cell death control in Arabidopsis disease resistance. Biotechnol Prog 20 426–442 [DOI] [PubMed] [Google Scholar]
- Albert I, Albert R (2004) Conserved network motifs allow protein-protein interaction prediction. Bioinformatics 20 3346–3352 [DOI] [PubMed] [Google Scholar]
- Albert I, Thakar J, Li S, Zhang R, Albert R (2008) Boolean network simulations for life scientists. Source Code Biol Med 3 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert R, DasGupta B, Dondi R, Kachalo S, Sontag E, Zelikovsky A, Westbrooks K (2007) A novel method for signal transduction network inference from indirect experimental evidence. J Comput Biol 14 927–949 [DOI] [PubMed] [Google Scholar]
- Albert R, Jeong H, Barabási AL (2000) Error and attack tolerance of complex networks. Nature 406 378–382 [DOI] [PubMed] [Google Scholar]
- Albert R, Wang RS (2009) Discrete dynamic modeling of cellular signaling networks. Methods Enzymol 467 281–306 [DOI] [PubMed] [Google Scholar]
- Ali R, Ma W, Lemtiri-Chlieh F, Tsaltas D, Leng Q, von Bodman S, Berkowitz GA (2007) Death don't have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 19 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GJ, Kuchitsu K, Chu SP, Murata Y, Schroeder JI (1999) Arabidopsis abi1-1 and abi2-1 phosphatase mutations reduce abscisic acid-induced cytoplasmic calcium rises in guard cells. Plant Cell 11 1785–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Ogata Y, Shibata D (2007) Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol 48 381–390 [DOI] [PubMed] [Google Scholar]
- Assmann SM, Albert R (2009) Discrete dynamic modeling with asynchronous update, or how to model complex systems in the absence of quantitative information. Methods Mol Biol 553 207–225 [DOI] [PubMed] [Google Scholar]
- Betts RA, Boucher O, Collins M, Cox PM, Falloon PD, Gedney N, Hemming DL, Huntingford C, Jones CD, Sexton DM, et al (2007) Projected increase in continental runoff due to plant responses to increasing carbon dioxide. Nature 448 1037–1041 [DOI] [PubMed] [Google Scholar]
- Bhalla US, Iyengar R (1999) Emergent properties of networks of biological signaling pathways. Science 283 381–387 [DOI] [PubMed] [Google Scholar]
- Bihler H, Eing C, Hebeisen S, Roller A, Czempinski K, Bertl A (2005) TPK1 is a vacuolar ion channel different from the slow-vacuolar cation channel. Plant Physiol 139 417–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright J, Desikan R, Hancock JT, Weir IS, Neill SJ (2006) ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J 45 113–122 [DOI] [PubMed] [Google Scholar]
- Cheong YH, Pandey GK, Grant JJ, Batistic O, Li L, Kim BG, Lee SC, Kudla J, Luan S (2007) Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J 52 223–239 [DOI] [PubMed] [Google Scholar]
- Cho D, Kim SA, Murata Y, Lee S, Jae SK, Nam HG, Kwak JM (2009) De-regulated expression of the plant glutamate receptor homolog AtGLR3.1 impairs long-term Ca2+-programmed stomatal closure. Plant J 58 437–449 [DOI] [PubMed] [Google Scholar]
- Christie JM, Reymond P, Powell GK, Bernasconi P, Raibekas AA, Liscum E, Briggs WR (1998) Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science 282 1698–1701 [DOI] [PubMed] [Google Scholar]
- Colcombet J, Hirt H (2008) Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J 413 217–226 [DOI] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C (2005) A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol 15 1196–1200 [DOI] [PubMed] [Google Scholar]
- Ding Z, Li S, An X, Liu X, Qin H, Wang D (2009) Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J Genet Genomics 36 17–29 [DOI] [PubMed] [Google Scholar]
- Eungdamrong NJ, Iyengar R (2004) Computational approaches for modeling regulatory cellular networks. Trends Cell Biol 14 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan LM, Zhang W, Chen JG, Taylor JP, Jones AM, Assmann SM (2008) Abscisic acid regulation of guard-cell K+ and anion channels in Gβ- and RGS-deficient Arabidopsis lines. Proc Natl Acad Sci USA 105 8476–8481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, et al (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA 106 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert A, Isayenkov S, Voelker C, Czempinski K, Maathuis FJ (2007) The two-pore channel TPK1 gene encodes the vacuolar K+ conductance and plays a role in K+ homeostasis. Proc Natl Acad Sci USA 104 10726–10731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonugunta VK, Srivastava N, Puli MR, Raghavendra AS (2008) Nitric oxide production occurs after cytosolic alkalinization during stomatal closure induced by abscisic acid. Plant Cell Environ 31 1717–1724 [DOI] [PubMed] [Google Scholar]
- He X, Zhang J (2006) Why do hubs tend to be essential in protein networks? PLoS Genet 2 e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosy E, Vavasseur A, Mouline K, Dreyer I, Gaymard F, Poree F, Boucherez J, Lebaudy A, Bouchez D, Very AA, et al (2003) The Arabidopsis outward K+ channel GORK is involved in regulation of stomatal movements and plant transpiration. Proc Natl Acad Sci USA 100 5549–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MM, Tani C, Watanabe-Sugimoto M, Uraji M, Jahan MS, Masuda C, Nakamura Y, Mori IC, Murata Y (2009) Myrosinases, TGG1 and TGG2, redundantly function in ABA and MeJA signaling in Arabidopsis guard cells. Plant Cell Physiol 50 1171–1175 [DOI] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabási AL, Oltvai ZN (2001) Lethality and centrality in protein networks. Nature 411 41–42 [DOI] [PubMed] [Google Scholar]
- Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL (2000) The large-scale organization of metabolic networks. Nature 407 651–654 [DOI] [PubMed] [Google Scholar]
- Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, Nahm BH, Choi YD, Cheong JJ (2008) Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol 146 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachalo S, Zhang R, Sontag E, Albert R, DasGupta B (2008) NET-SYNTHESIS: a software for synthesis, inference and simplification of signal transduction networks. Bioinformatics 24 293–295 [DOI] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K (2001) Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414 656–660 [DOI] [PubMed] [Google Scholar]
- Kirschbaum MU (2004) Direct and indirect climate change effects on photosynthesis and transpiration. Plant Biol (Stuttg) 6 242–253 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM (1984) The isolation and characterization of abscisic acid insensitive mutants of Arabidopsis thaliana. Physiol Plant 61 377–383 [Google Scholar]
- Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI (2006) The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol 140 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde S, Ehrhardt DW, Loque D, Chen J, Rhee SY, Frommer WB (2008) Molecular and cellular approaches for the detection of protein-protein interactions: latest techniques and current limitations. Plant J 53 610–635 [DOI] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S (2009) A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA 106 21419–21424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Assmann SM, Albert R (2006) Predicting essential components of signal transduction networks: a dynamic model of guard cell abscisic acid signaling. PLoS Biol 4 e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324 1064–1068 [DOI] [PubMed] [Google Scholar]
- Meinhard M, Grill E (2001) Hydrogen peroxide is a regulator of ABI1, a protein phosphatase 2C from Arabidopsis. FEBS Lett 508 443–446 [DOI] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25 295–303 [DOI] [PubMed] [Google Scholar]
- Merlot S, Leonhardt N, Fenzi F, Valon C, Costa M, Piette L, Vavasseur A, Genty B, Boivin K, Muller A, et al (2007) Constitutive activation of a plasma membrane H+-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J 26 3216–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Mustilli AC, Genty B, North H, Lefebvre V, Sotta B, Vavasseur A, Giraudat J (2002) Use of infrared thermal imaging to isolate Arabidopsis mutants defective in stomatal regulation. Plant J 30 601–609 [DOI] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264 1452–1455 [DOI] [PubMed] [Google Scholar]
- Mori IC, Murata Y, Yang Y, Munemasa S, Wang YF, Andreoli S, Tiriac H, Alonso JM, Harper JF, Ecker JR, et al (2006) CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+-permeable channels and stomatal closure. PLoS Biol 4 e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y, Pei ZM, Mori IC, Schroeder J (2001) Abscisic acid activation of plasma membrane Ca2+ channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1-1 and abi2-1 protein phosphatase 2C mutants. Plant Cell 13 2513–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi J, Matsuda O, Nagasawa T, Oba Y, Takahashi H, Kawai-Yamada M, Uchimiya H, Hashimoto M, Iba K (2008) CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452 483–486 [DOI] [PubMed] [Google Scholar]
- Osakabe Y, Maruyama K, Seki M, Satou M, Shinozaki K, Yamaguchi-Shinozaki K (2005) Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell 17 1105–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Nelson DC, Assmann SM (2009) Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136 136–148 [DOI] [PubMed] [Google Scholar]
- Pandey S, Zhang W, Assmann SM (2007) Roles of ion channels and transporters in guard cell signal transduction. FEBS Lett 581 2325–2336 [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei ZM, Kuchitsu K, Ward JM, Schwarz M, Schroeder JI (1997) Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell 9 409–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera IY, Hung CY, Moore CD, Stevenson-Paulik J, Boss WF (2008) Transgenic Arabidopsis plants expressing the type 1 inositol 5-phosphatase exhibit increased drought tolerance and altered abscisic acid signaling. Plant Cell 20 2876–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL (2004) Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J 37 354–369 [DOI] [PubMed] [Google Scholar]
- Saez A, Robert N, Maktabi MH, Schroeder JI, Serrano R, Rodriguez PL (2006) Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol 141 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito N, Munemasa S, Nakamura Y, Shimoishi Y, Mori IC, Murata Y (2008) Roles of RCN1, regulatory A subunit of protein phosphatase 2A, in methyl jasmonate signaling and signal crosstalk between methyl jasmonate and abscisic acid. Plant Cell Physiol 49 1396–1401 [DOI] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Marquez JA (2009. a) The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462 665–668 [DOI] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Marquez JA, Cutler SR, Rodriguez PL (2009. b) Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J 60 575–588 [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Last RL, Pichersky E (2008) Harnessing plant trichome biochemistry for the production of useful compounds. Plant J 54 702–711 [DOI] [PubMed] [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, Alon U (2002) Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet 31 64–68 [DOI] [PubMed] [Google Scholar]
- Sirichandra C, Gu D, Hu HC, Davanture M, Lee S, Djaoui M, Valot B, Zivy M, Leung J, Merlot S, et al (2009) Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett 583 2982–2986 [DOI] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA 106 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T, Kollist H, Wang YF, Nishimura N, Chan WY, Valerio G, Lamminmaki A, Brosche M, Moldau H, Desikan R, et al (2008) SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Holroyd G, Hetherington AM, Ng CK (2004) Seeing ‘cool’ and ‘hot’: infrared thermography as a tool for non-invasive, high-throughput screening of Arabidopsis guard cell signalling mutants. J Exp Bot 55 1187–1193 [DOI] [PubMed] [Google Scholar]
- Ward JM, Maser P, Schroeder JI (2009) Plant ion channels: gene families, physiology, and functional genomics analyses. Annu Rev Physiol 71 59–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM (1994) The ABCs of floral homeotic genes. Cell 78 203–209 [DOI] [PubMed] [Google Scholar]
- Worrall D, Liang YK, Alvarez S, Holroyd GH, Spiegel S, Panagopulos M, Gray JE, Hetherington AM (2008) Involvement of sphingosine kinase in plant cell signalling. Plant J 56 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Wang Y, Williamson L, Holroyd GH, Tagliavia C, Murchie E, Theobald J, Knight MR, Davies WJ, Leyser HM, et al (2006) The identification of genes involved in the stomatal response to reduced atmospheric relative humidity. Curr Biol 16 882–887 [DOI] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH (2006) A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125 1347–1360 [DOI] [PubMed] [Google Scholar]
- Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI (2008) Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K (2006) The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem 281 5310–5318 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhu H, Zhang Q, Li M, Yan M, Wang R, Wang L, Welti R, Zhang W, Wang X (2009) Phospholipase Dα1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 21 2357–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Zhang W, Stanley BA, Assmann SM (2008) Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. Plant Cell 20 3210–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Dai S, McClung S, Yan X, Chen S (2009) Functional differentiation of Brassica napus guard cells and mesophyll cells revealed by comparative proteomics. Mol Cell Proteomics 8 752–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SY, Yu XC, Wang XJ, Zhao R, Li Y, Fan RC, Shang Y, Du SY, Wang XF, Wu FQ, et al (2007) Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19 3019–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]