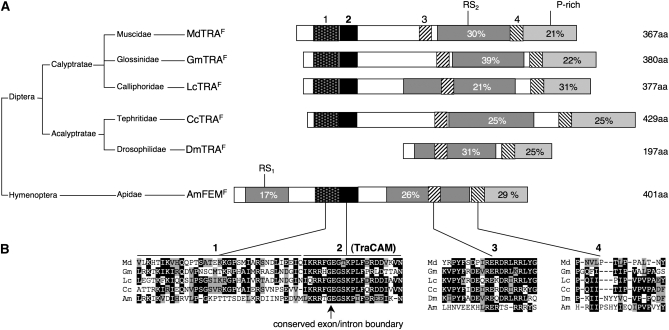
Figure 3.—
Small conserved protein motifs in TRA homologs. (A) Schematic alignment of proteins encoded by the tra ortholog of Musca domestica (MdTRAF), Glossina morsitans (GmTRAF), Lucilia cuprina (LcTRAF), Ceratitis capitata (CcTRAF), Drosophila melanogaster (DmTRAF), and Apis mellifera (AmFEMF). The percentage of arginine/serine residues in the RS domains (dark shading) and the percentage of proline residues in the P-rich domains (light shading) are indicated within the boxes. The relative position and size of the four conserved motifs are indicated by differently shaded boxes (B) Sequence alignment of these small motifs. The conserved exon/intron boundary in motif 2 (TRA-CAM) is indicated by an arrow. In Musca, the glutamine (Q) directly upstream of the conserved TRA-CAM domain is replaced by an arginine (R) in the dominant allele MdtraD; see also Figure 7A and text.